Cancer Data Science Pulse
FireCloud: A Secure Platform For Data Analysis Powered by Terra
Here, we’re spotlighting FireCloud, another NCI resource for accessing data, running analysis, and collaborating with others in the cancer research community. FireCloud is a Broad Institute project funded by NCI to empower cancer researchers to access data, run analysis tools, and collaborate securely in the cloud. Powered by Terra, FireCloud is a secure, scalable, cloud-native platform developed by the Broad Institute, Microsoft, and Verily, an Alphabet company.
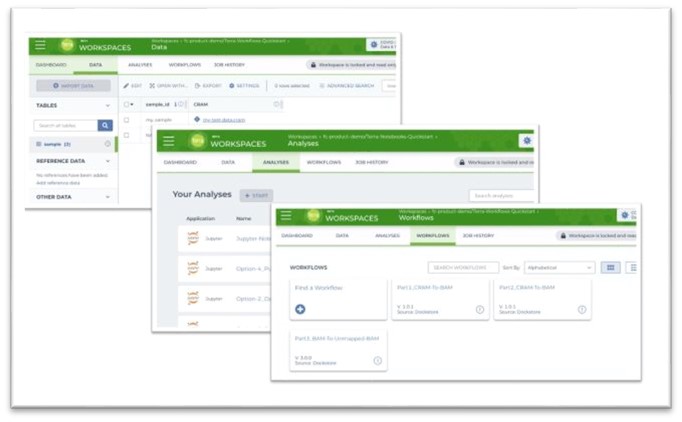
Working within FireCloud centers on workspaces that serve as computational “sandboxes” for bringing together data and tools. Workspaces also provide a security perimeter for controlling who can access, edit, and compute within a collaboration. Each workspace is associated with a dedicated storage space that researchers can use to store their private data and the outputs of their analyses. The workspace interface consists of a dashboard that summarizes information about the workspace and three additional tabs:
- Data: For importing researchers’ proprietary data, accessing FireCloud hosted data or other cloud-hosted data, and keeping it all organized
- Analysis: For configuring and launching interactive analytical applications
- Workflows: For configuring, launching, and monitoring execution of automated workflows
Workspaces Deliver Study Reproducibility and Secure Sharing
Researchers can share their workspaces across institutional boundaries, making it easy to collaborate with colleagues across the street or around the world. To protect the confidentiality of controlled-access data, the platform includes strong security measures to ensure access is not inadvertently shared with unauthorized users.
FireCloud users can make their workspace fully public to serve as a companion resource for a publication, as described in a blog post. This enables others in the research community to reproduce the published work and reuse the analysis methods in their own work. For studies involving controlled-access data, researchers should instead create a version of their workspace that demonstrates usage of the analysis methods on open-access data as a substitute.
Data: Accessing, Importing, Organizing, and Storing
FireCloud provides access to many large NCI-funded data sets, including TARGET and TCGA. The underlying data import system allows researchers to pull together data from many sources —whether hosted in FireCloud or in a connected third-party repository— to perform federated analysis without having to download or store copies of the data themselves.
Researchers can upload their own proprietary data within their secure workspace. This upload makes it possible to put their data and analysis findings in context with larger study cohorts, gain statistical power, and leverage complementary data types to interrogate complex biological mechanisms.
Workflows: User-friendly and Scalable
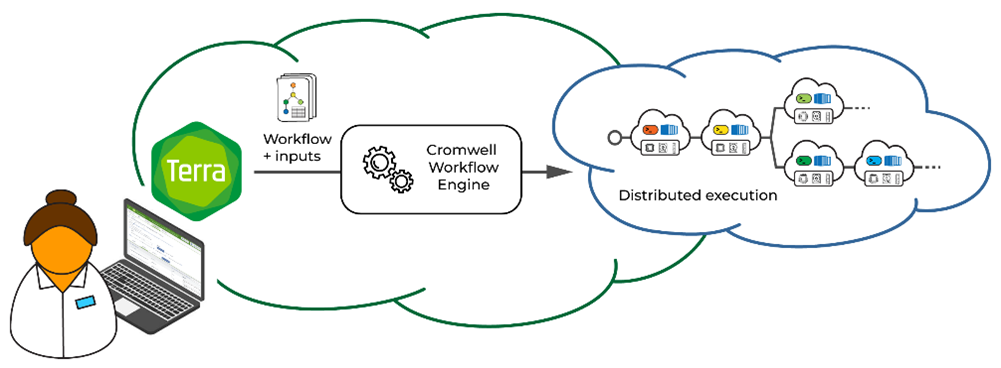
Analyses that involve large amounts of data and multiple compute-intensive and/or repetitive operations greatly benefit from being automated using workflow scripts. FireCloud, through Terra, includes a built-in Cromwell server that is able to run workflows written in the Workflow Description Language (WDL), an open standard stewarded by the OpenWDL community. A WDL workflow describes a series of data processing steps—comprising individual tool command lines—chained together into a pipeline.
For each step in the workflow, the workflow code points to a tool container in a registry such as Dockerhub or the Google Container Repository. These registries allow developers to upload and share their command-line tools in containers that bundle all necessary software dependencies, such as third-party libraries.
Researchers can submit their WDL workflow to the Cromwell server, which interprets the workflow code and generates batches of individual analysis jobs corresponding to the steps in the workflow. The server then dispatches these jobs to a Google Cloud service that handles their execution on virtual machines in the cloud using the specified tool containers.
A key feature of this system is its scalability. Workloads can be distributed across large numbers of virtual machines, making it possible to parallelize operations and scale data processing to thousands of inputs, without requiring any special effort on the part of the user. In addition, the system includes features that enable robust recovery in case of error and support cloud optimizations to minimize cost and runtime. Combining scripts with containerized tools in this way helps ensure the highest level of reproducibility. Using the FireCloud platform, computational biologists can distribute pipelines to collaborators (or the general community) so future analysis is done the same way. Also, as a result, developers can encapsulate very complex analyses to make them more accessible for novice users, while remaining flexible for further development.
Researchers who do not want to develop their own workflows can take advantage of a growing collection of community developed WDL workflows available in public repositories. For example, Dockstore holds high-quality workflows contributed by various labs and consortia covering numerous use cases, including germline and somatic variant discovery, single-cell data processing, viral genomics, and more.
Analysis: Tailored or Out-of-the Box Tools
Cancer researchers can also take advantage of FireCloud’s flexible environment to explore and analyze data interactively using popular applications, such as RStudio, Jupyter Notebooks, and Galaxy. Jupyter Notebooks and Galaxy are particularly suitable for researchers who have not previously received computational training. The FireCloud and Terra documentation includes detailed tutorials and videos that enable novices to get started quickly and to progress at their own pace.
The system also provides pre-built environments that include popular tool suites, such as Bioconductor, that work out of the box, as well as the ability to install any additional command-line tools and packages as needed. The system also allows researchers to customize their environment by specifying their own container and/or an installation script which can be used to ensure reproducibility of an analysis when sharing code with collaborators.
In terms of computing power, researchers can choose to use a default hardware configuration that is designed to serve as a user-friendly "one size fits most" option. Alternatively, they can choose to customize the configuration to control how much computing power they want to mobilize, and to use advanced hardware such as GPUs.
Case Study: Reproducible Workflows through PANOPLY
The PANOPLY proteogenomics analysis framework re-implements methods from flagship Clinical Proteomic Tumor Analysis Consortium studies (Mani et al., 2021). This framework consists of a set of reproducible workflows intended for reuse by the research community. You can see the workflows in action in a recent research publication that used PANOPLY to characterize the proteogenomics of lung squamous cell carcinoma (Satpathy et al., 2021). The developers of PANOPLY explicitly designed their framework to take advantage of FireCloud's capabilities.
A public tutorial workspace demonstrates how to apply these methods to perform a proteogenomics analysis of data from BRCA samples (Krug et al., 2020) and a lung adenocarcinoma (LUAD) cohort (Gillette et al., 2020), which are accessible through the FireCloud Data Library.
Anyone can view this tutorial workspace dashboard (which includes usage instructions) without login; logging into a free account (noted below) provides access to the full workspace contents.
Ready to Get Started?
You can find tutorial workspaces covering other cancer analysis use cases in the FireCloud/Terra Showcase. If you have questions about setting up a free account or using any of these tools, please reach out to the support team through the help desk or the Terra Community Forum.
Read the other blogs in the series:
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (40)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (25)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (13)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

![Screenshot of the PANOPLY public tutorial interface that reads, ABOUT THE WORKSPACE PANOPLY Tutorial
PANOPLY is a platform for applying state-of-the-art statistical and machine learning algorithms to transform multi-omic data from cancer samples into biologically meaningful and interpretable results. PANOPLY leverages Terra-a cloud-native platform for extreme-scale data analysis, sharing, and collaboration-to host proteogenomic workflows, and is designed to be flexible, automated, reproducible, scalable, and secure. A wide array of algorithms applicable to all cancer types have been implemented, and we highlight the application of PANOPLY to the analysis of cancer proteogenomic data.
This PANOPLY tutorial provides a tour of how to use the PANOPLY proteogenomic data analysis pipeline, using the breast cancer dataset published in Mertins, et. al.] The input dataset ( tutorial-brca-input.zip file) can be found in the tutorial subdirectory, along with a HTML version of this tutorial.](/sites/default/files/2022-12/FireCloudPANOPLYWorkspace.png)
Leave a Reply