Cancer Data Science Pulse
The Quest for Harmonized Data
A big challenge in making data usable for analysis is that not all data annotations conform to the same standard. Hundreds of studies are yielding high-quality data, but each study may use a different data model, data elements, or annotations. Such inconsistency makes it nearly impossible to compare these findings. Indeed, researchers often state this is the primary reason they do not use data from other studies, as it takes too long to clean individual data sets to make them compatible.
Researchers have fought for quality data standards for years. The FAIR principles (i.e., data that are Findable, Accessible, Interoperable, and Reusable) were put in place to improve our ability to find and reuse data. The ultimate goal is to harmonize and aggregate data by applying certain uniform standards. If those standards are met prior to submission, it should make it easier to find the data (either by manual searches or by computer); to access data (with proper authentication and/or authorization); and to integrate data for analysis, storage, and processing.
The best model for harmonization would be easily adapted and simple to use. It would set a standard for how we describe the data. With such standards, our ability to compare the full range of data sets would increase exponentially.
Yet opinions differ on what constitutes the “right” model, as do choices of “standards.” If anything, there are too many standards, making it difficult to select the best. Even if the same general standard is followed, science changes over time, and small advances in methodology can mean big differences when comparing tomorrow’s studies with those from a decade ago. Add to this the variations in the types of data available—genomic, proteomic, metabolomic, histologic, imaging—and it’s clear to see why harmonization remains elusive.
Building a Better Mousetrap
This could soon change. Our quest to harmonize data has ushered in a new way of thinking about standardization. Now, rather than expecting everyone to adopt a particular model or standard, we’re seeking to leverage technology that can do some of this work for us, seamlessly and efficiently, with little if any manual intervention. Ideally, such harmonization also could be conducted retrospectively, after the data already have been collected, as a way of including older, legacy data sets.
NCI’s Center for Biomedical Informatics and Information Technology (CBIIT) has made groundbreaking advances in harmonization, from the early work in semantics and the development of the NCI Metathesaurus, Thesaurus, and cancer Data Standards Registry and Repository (caDSR), to the recent launch of the Center for Cancer Data Harmonization. The next step in the quest for harmonization can be found in the NCI Cancer MoonshotSM, which is leading the Metadata Automation DREAM Challenge. This challenge addresses, in particular, the time-consuming task of finding standardized metadata annotations for structured data. Such harmonization is vital to advance the usability of NCI’s Cancer Research Data Commons (CRDC).
The DREAM Challenge was designed to build a better mousetrap, or tool, to make aggregating and mapping data to the correct lexicon of terms and metadata a nearly seamless step for researchers. Such a tool would operate on individual datasets. Its main task would be to automatically scan data upon submission and align the fields and entries with standards. We think that having such a tool on hand would enable downstream processing by other tools to help further map and transform data, such as:
- Validating the data against the standards for any data errors
- Allowing the user to select (perhaps through drag and drop menus) different fields to create a customized format or to choose from an existing format
- Automatically mapping the data to the new format, as needed
- Identifying missing or dropped data
- Organizing and providing the new data in a format most useful for analyses
In short, we would have the ability to streamline both the submission of new data and the retrospective harmonization of older data. This would thus allow researchers to more easily access, analyze, interpret, and share their findings.
The Challenge is Underway
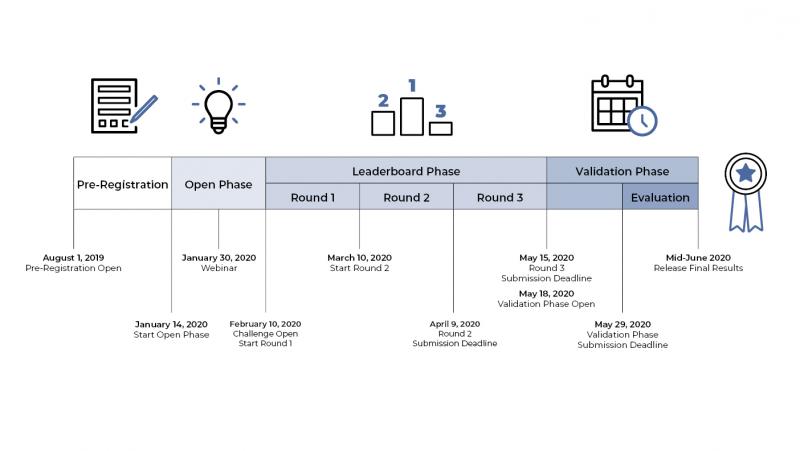
The DREAM Challenge launched on February 10, 2020. More than 100 participants have answered the call from across the United States and from 11 countries, such as Greece, Turkey, Korea, Australia, and Ireland. These competitors reflect diverse backgrounds and represent biotech companies and academia, as well as independent contractors.
Participants were given a step-by-step guide, structured biomedical data, and resources from the caDSR and the NCI Thesaurus. Using those files, Challenge competitors are working to develop solutions for automating annotation to describe the meaning of data fields and values. Such a system is expected to be generalizable and to work with different metadata and terminological resources. Challenge participants also are being encouraged to be creative in leveraging other external resources. They should, however, look for solutions that will use the metadata in the caDSR and supporting terminology from the NCI Thesaurus.
The participants will offer algorithms specifically designed to meet the needs of this Challenge. In the future, those algorithms would be built into libraries or incorporated into custom-built tools. In the end, the intent is to develop an approach for managing the diverse annotation requirements of the CRDC. Ideally, the teams will offer a way to support the annotation of data in flat files (headers, row values) according to their semantics—i.e., using identifiers for common data elements and concepts.
A successful Challenge will give us a scalable and faster mechanism for harmonizing input data to each CRDC repository’s standards, along with the flexibility to allow for new developments in the field. Furthermore, successful harmonization will give us a way to match fields/values with semantic concepts to make data useful to a broad range of applications—from input of a single data set to synchronization of legacy data in large existing repositories.
You can check the leaderboard for updates on how the Challenge is progressing. The competition is slated to close on May 29, 2020 and prizes will be announced in mid-June.
Leave a Reply
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (39)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (24)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (12)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

Robert Martin on April 30, 2020 at 04:10 p.m.