Cancer Data Science Pulse
Cancer Data Aggregator—The Engine that Could . . . Drive Data Aggregation in a Whole New Way
Cancer initiation and progression require complex interactions within the cellular environment that lead to uncontrollable cell growth. The National Cancer Institute (NCI) supports many complex studies, such as Human Tumor Cell Atlas Network (HTAN) and Clinical Proteomic Tumor Analysis Consortium (CPTAC), to increase our knowledge of cancer to ultimately advance toward better prevention and treatments options. These studies generate a wealth of diverse data sets that require aggregation to enable researchers to combine information for meaningful interpretation. However, the integration of these data is currently hampered by the fact that data are stored in separate repositories, making aggregation difficult.
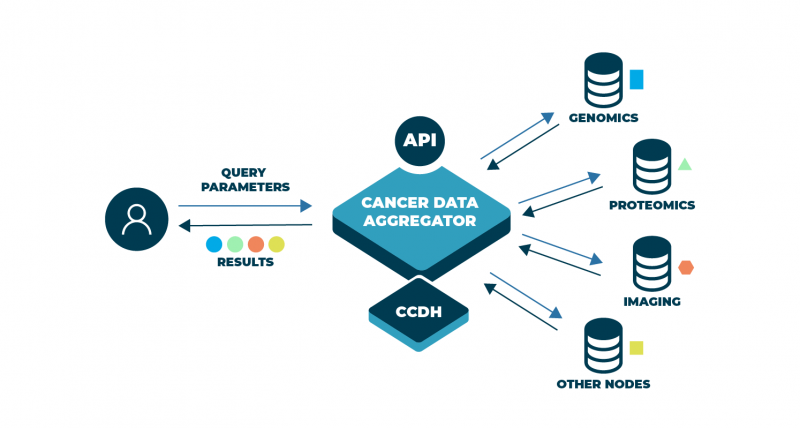
The need for an integrative approach is the basis underlying the next important step in the development of the NCI Cancer Research Data Commons (CRDC) and the tools that support it. A new Cancer Data Aggregator (CDA) is being designed and developed to allow scientists to crosstalk among very diverse data sets—with information from the fields of genomics, proteomics, single-cell, radiology, molecular imaging, clinical findings, and more. Furthermore, it will facilitate interoperability within the larger data ecosystem by connecting to additional data repositories outside of NCI’s CRDC.
Today, researchers often are able to analyze harmonized data from a single repository, depending on how well those data have followed a certain standard when submitted. What’s missing, though, is a way to search and collect information across the CRDC by making use of a common (or overarching) model developed by the Center for Cancer Data Harmonization (CCDH). This model would allow us to compare not only similar data but also highly diverse information, ranging from DNA to demographics, on a scalable, consistent basis.
Within NCI’s CRDC there are repositories of data representing genomics, proteomics, imaging, histopathology, and other clinical features, as well as information on the patients themselves. Using multi-omics technology, researchers can unravel information about genes and proteins to look for clues as to why and how cancer develops and identify targets for treatments. When blended, this information offers infinite possibilities for research, as the sum of these data will be exceedingly greater than the parts alone.
Ironically what makes the CRDC so unique—the diversity of the data—is also what makes searching and comparing those data so difficult. Today scientists are able to use queries to sift through information on demographics, lab tests, treatment regimens, physiological monitoring results, and more. For genomics, a single query can return results from millions of sequences to pinpoint areas of the genetic code that are most relevant. Queries into proteomics allow us to see how proteins are structured, function, and interact to regulate our body’s systems. But each of these queries yields results only within those particular types of data—clinical, genomics, proteomics. To aggregate data across the CRDC (and other repositories), we need to develop a way to reach into many different data sets and extract only that information which is needed the most.
An aggregator could drive this research further—perhaps through the use of a single question that would query all the data sets at one time. For example, with the CDA an investigator could integrate findings on a specific mutation in a gene related to a type of cancer, link it to a protein or pathway to reveal the mechanism of action, and tie those findings to highly specific patient characteristics, such as age, tumor grade, and type of treatment. The results could help develop a model to precisely predict how effective a certain medication or therapy will be based on a patient’s molecular characteristics before it’s ever administered. Such integrated research would allow clinicians to target treatment that will give their patients the best outcomes and in the shortest time.
Although the specific functionality is still in the early phases of planning and development, the CDA will be built to include:
- A centralized repository for basic clinical and biospecimen metadata with standardized terms used to describe data within the various repositories (to make the data discoverable); and
- A search engine to enable CRDC users to submit queries across a wide variety of patient data and experimental criteria.
Work on the CDA is just getting underway. The contract to develop this new tool was awarded through Leidos Biomedical Research, Inc., on May 6th, 2020, to a consortium led by the Broad Institute. Other consortia members include the Institute for Systems Biology, Seven Bridges, and General Dynamics Information Technology, Inc. This coalition has been charged with creating an aggregator tool that is capable of searching and aggregating multi-modal data. With the CDA, scientists will be able to reach deep into diverse data sets and extract meaningful results to further advance scientific discovery.
The CDA launch is planned in 2021. For more information, read the news article announcing the CDA award.
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (39)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (24)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (12)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

Leave a Reply