Cancer Data Science Pulse
Imaging Data Commons Brings the Power of the Cloud to Cancer Research
Today we have an array of high-tech imaging tools for diagnosing and tracking cancer. Images from digital microscopy, Computed Tomography (CT), Positron Emission Tomography (PET), Magnetic Resonance Imaging (MRI), ultrasound, and X-ray give an inside look at how cancer develops and progresses. Such noninvasive images also allow clinicians to screen for and diagnose cancer, even in patients who don’t show outward signs of disease.
These images have been generated and collected for decades, but on the whole, have not been available for widespread use by researchers. Now, with recent innovations in imaging AI, there’s been an even greater surge of interest in using imaging content for scientific discovery. The Human Tumor Atlas Network (HTAN) is one example. This NCI-funded Cancer MoonshotSM initiative has been charged with mapping the dynamic cellular, morphological, and molecular features of human cancers as they evolve from precancerous lesions to advanced disease.
As more medical images are amassed, many are being stored at academic centers and collected by repositories around the globe, including The Cancer Imaging Archive (TCIA). These ever-burgeoning repositories offer imaging data for secondary analysis and the development and validation of software tools. What’s been lacking, however, is a way to easily search, sort, and create data cohorts. Once those cohorts are collected, we also need a way to connect to other data elements, like genomics, and perform analysis on a grand scale, such as within a cloud-based infrastructure. With all these pieces in place we have a greater opportunity to further advance cancer research.
Imaging Data Commons
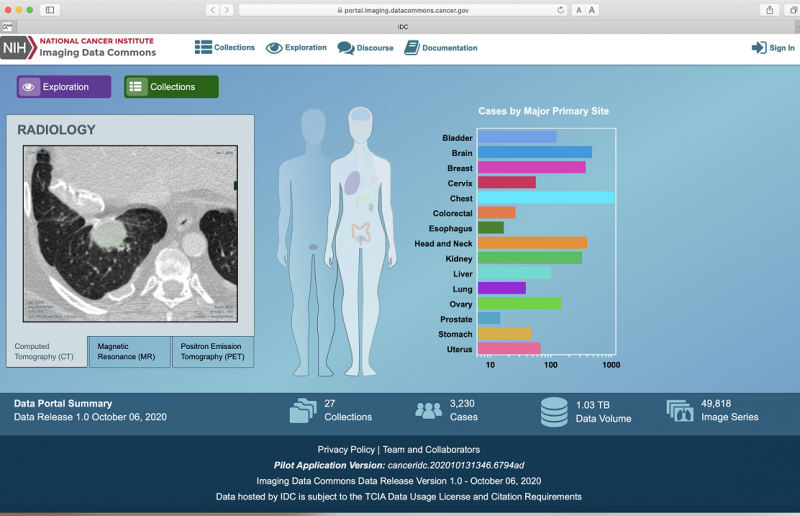
On October 20, 2020, NCI launched the Imaging Data Commons (IDC), the latest data repository to be offered within the Cancer Research Data Commons (CRDC) infrastructure. The IDC was developed in partnership with the Frederick National Laboratory for Cancer Research and the Brigham and Women’s Hospital, Harvard Medical School, with Drs. Ron Kikinis and Andrey Fedorov as co-principal investigators; and with support from the team at the Institute for Systems Biology, led by William Longabaugh.
Through the IDC, both researchers and clinicians will have access to a wide range of cancer-related images, including radiology and pathology imaging data, as well as their accompanying metadata. The IDC also includes tools for searching, identifying, and viewing images and for creating image cohorts to allow for further analysis in the cloud using the NCI’s Cancer Cloud Resources.
As a centralized resource for imaging data, IDC will offer documented provenance, search and visualization capabilities, harmonization, standardization, and quality control. Such measures will ensure that the data adhere to unified standards of the field and the FAIR principle of making data Findable, Accessible, Interoperable, and Reusable.
Most importantly, as part of the larger CRDC, this new imaging repository is cloud-based. This platform gives researchers an efficient way of locating and using image analysis software tools; connecting imaging data with findings from other fields, such as genomics and proteomics; and performing computation that draws on the elastic capabilities of cloud compute, allowing researchers to create workspaces in NCI’s Cancer Cloud Resources to perform their work from any location and with minimal local resources.
The First Release
With this first release, IDC offers a variety of images, including CT, MRI, and ultrasound, collected within the TCIA. IDC images adhere to the Digital Imaging and Communications in Medicine (or DICOM) standards, an internationally recognized standard for acquisition and electronic communication of medical images. This ensures that images can be easily compared, even when they are obtained from medical devices from multiple manufacturers.
Over the coming months we also will add digital pathology collections from TCIA. A DICOM standard currently exists for digital pathology, although it has not been universally adopted by all device manufacturers. Because digital pathology images in TCIA currently are stored in vendor-specific formats, part of IDC’s tasks will be to ensure these existing images meet DICOM pathology standards prior to making them available to researchers.
These measures to standardize images should help, but even content that meets the minimum DICOM criteria may lack vital metadata information. Metadata should, at the very least, include physician annotations and tumor segmentations, which are important for in-depth or cross-study comparison.
Thus, we also are working with NCI’s Center for Cancer Data Harmonization (CCDH) to further harmonize metadata and models both within and beyond IDC so the images can be used across the CRDC. This will facilitate the comparison of data within each data repository, as well as across the full data infrastructure. Searching and comparing data also will be more efficient with the advent of a new Cancer Data Aggregator, a tool that is now in early development.
The IDC website has an easy-to-navigate process for researchers who wish to search, view, and analyze images in the cloud. Researchers also can use this resource to develop new AI tools to better understand how cancers occur and how they might be treated. Through the IDC, we hope to empower researchers to conduct new studies that are fully integrative, rigorous, easily traceable, and which can be compared across diverse studies for more comprehensive results than ever before.
Leave a Reply
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (40)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (25)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (13)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

Dafeng JI on October 21, 2020 at 08:04 p.m.