Cancer Data Science Pulse
Using the Cloud for Cancer Research—Overcoming Common Barriers
The cloud offers a wide range of repositories, tools, and resources for advancing your cancer research, but it might not be for everyone. If you’re not using the cloud, what’s standing in your way?
Below are three common reasons you might be avoiding the cloud, along with some tips and an example to help you address these barriers. Who knows; moving to the cloud may help transform your research in ways you haven’t considered before.
What’s Keeping You from Using the Cloud?
Barrier #1: It’s too expensive.
It’s true that long hours spent online can add up quickly in fees. However, there are ways around these costs.
Cloud computing operates under a “pay as you go” model, with settings that automatically shut off unused resources to save costs. You’re only charged for the resources you need, when you need them. Consider a test drive first, which will allow you to sample the service before you buy. NCI’s Cancer Research Data Commons offers new users up to $300 in computation and storage credits.
NCI-funded cloud resources (i.e., Broad Institute FireCloud, Institute for Systems Biology Cancer Gateway in the Cloud, and Seven Bridges Cancer Genomics Cloud, powered by Velsera) also offer tools to help you manage costs. For example, if you’re working with genomics data and want to use Amazon Web Services tools, Seven Bridges offers tools that feature built-in compute cost discounts. You also can use the Cost Estimator to see the cost of execution before running an analysis. Visit the Cloud Infrastructure Pricing webpage for information on how costs are incurred.
Another approach to keeping costs in check is to develop your workflow locally, on a small scale, before moving it to the cloud for larger and more sophisticated data analyses. This enables you to work out any “bugs” before you move to the cloud environment, where slow-downs and troubleshooting become more costly.
Ask yourself, “Can you afford not to use it?” The cloud offers you ways to streamline and standardize your work (for easier replication), which could save you time and resources. Cloud computing also could lead to savings compared to institutional supercomputers, which can cost thousands of dollars per year for external users.
Barrier #2: It’s not secure.
Any time data leaves your desktop you worry about security. NCI understands that fear, and so we follow industry best practices with regard to access control, network security, and enacting a regularly updated and modernized system.
In addition, the cloud resource teams that help manage NCI’s CRDC offer secure workspaces for users like you, including access to both open and controlled access data sets, as well as importing your own data.
Having these security measures in place lets you track your data so you can feel confident about the storage, sharing, and usage of those data.
Barrier #3: It would take time to transfer my work to the cloud.
If you’re studying the genetics underlying cancer or working with biomedical images, you know these data sets can be massive. Genomics, proteomics, and imaging data sets (e.g., data from The Cancer Genome Atlas [TCGA], the Clinical Proteomic Tumor Analysis Consortium [CPTAC], and NCI’s Imaging Data Commons) are difficult to store and analyze on local machines. Even on more robust academic ecosystems, you may find it takes a lot of time to process a query. You may find even running a simple analysis can overtax your local systems.
Cloud computing can help make your work more efficient. In addition, you’ll have access to tools and other resources that are only available in a cloud environment. You also can find pre-built (and fully tested) tools. Use these as stand-alone tools or incorporate them into your pipeline for added efficiency. For example, the cloud offers access to rich data from NCI’s CRDC, with more than 10 petabytes available. You can learn about these data using the Cancer Data Aggregator (CDA), a point-and-search tool that lets you collect, explore, and analyze data across NCI’s CRDC, including the:
- Genomic Data Commons,
- Imaging Data Commons,
- Proteomic Data Commons,
- Integrated Canine Data Commons,
- General Commons (formerly known as the Cancer Data Service), and
- Clinical and Translational Data Commons.
Prefer to bring your own data? You can access high-speed data tools to directly transfer your data from local environments. For example, Biowulf’s cgc-uploader on Helix lets you use parallelization, to give you a fast, efficient, and secure way of uploading data onto NCI’s Cancer Genomics Cloud.
See the Cloud in Action
Still not sure if the cloud is right for you? Below is a specific example to show how you might use the cloud for a small scale multi-modal analysis.
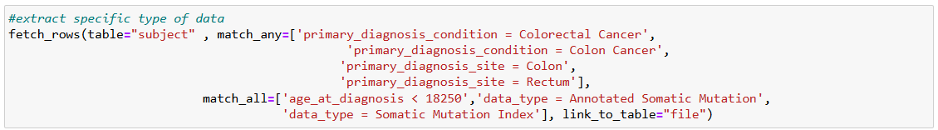
Suppose you want to explore the potential biological pathways associated with early-onset colorectal cancer (eCRC) by integrating multiple types of omics data. With the cloud resources from CRDC, you could begin by identifying patients with eCRC and normal-onset CRC using the CDA and the following query (see Figure 1). This will allow you to identify the appropriate genomic, proteomic, or RNA-sequencing data from the respective Data Commons.
You then can access the metadata in two ways:
- You can directly access the data through dbGaP, and download to your computer; or
- you can import from a DRS server directly in CGC environment.
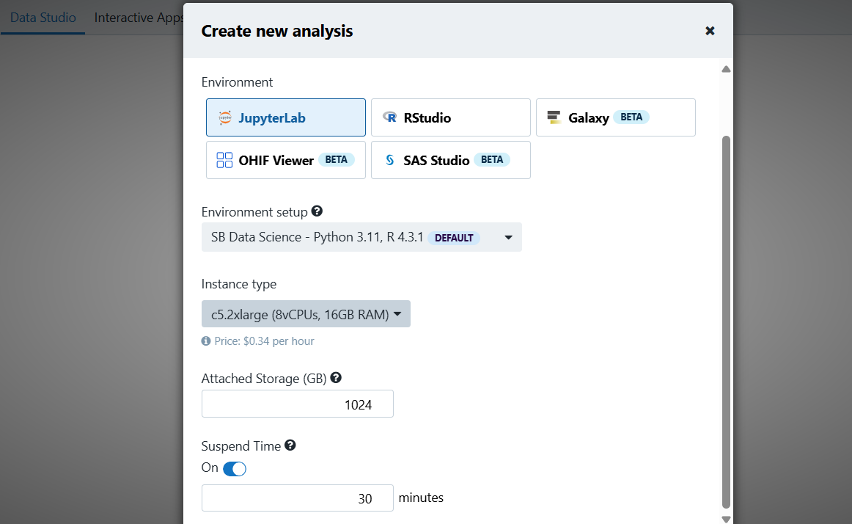
With these data in hand, you can explore a large inventory of public apps (more than 1,000 at the time of writing this blog post) in the Cancer Genomics Cloud (see Figure 2), or carry out the analysis using the Seven Bridges Data Studio, which very likely supports your favorite programming languages.
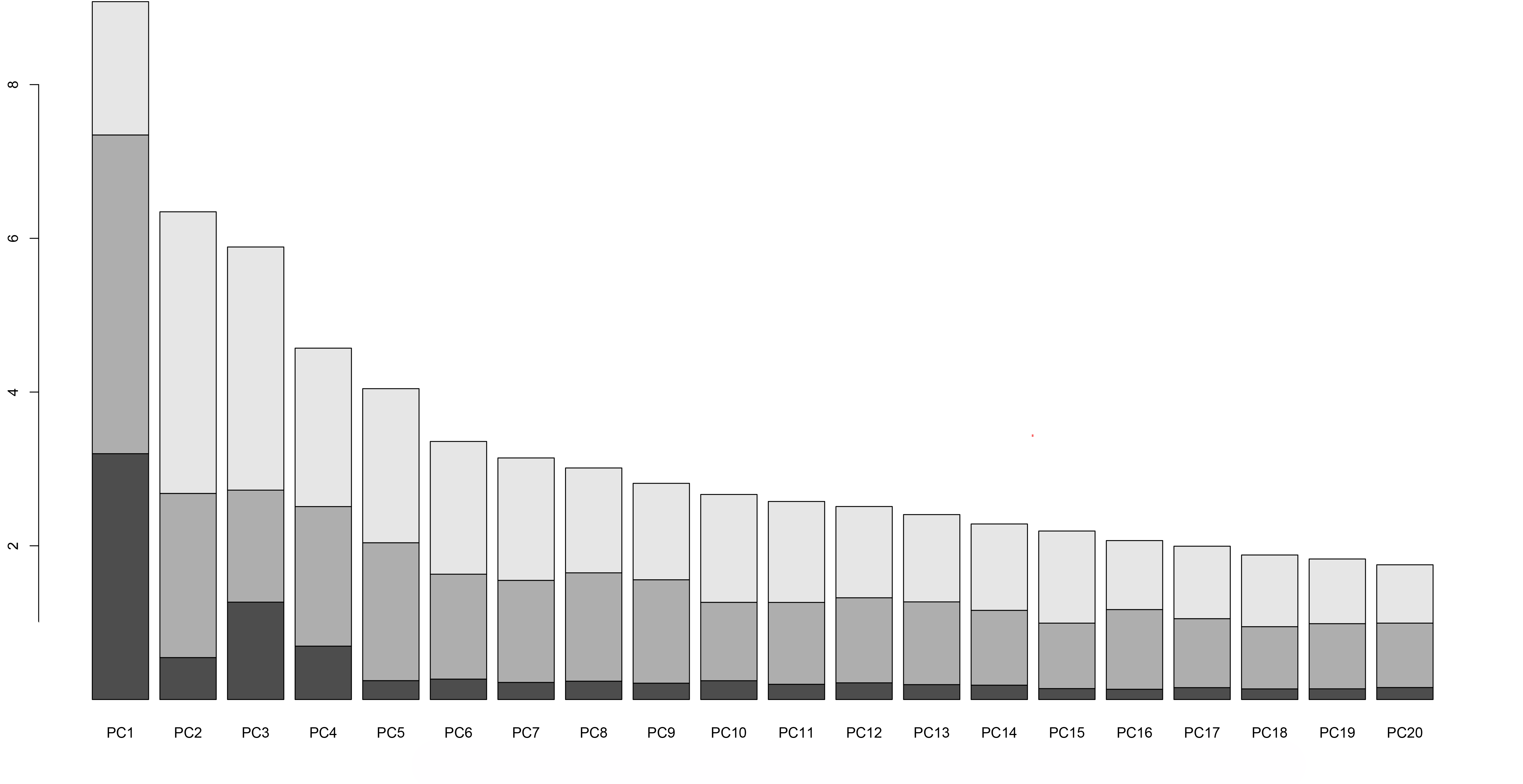
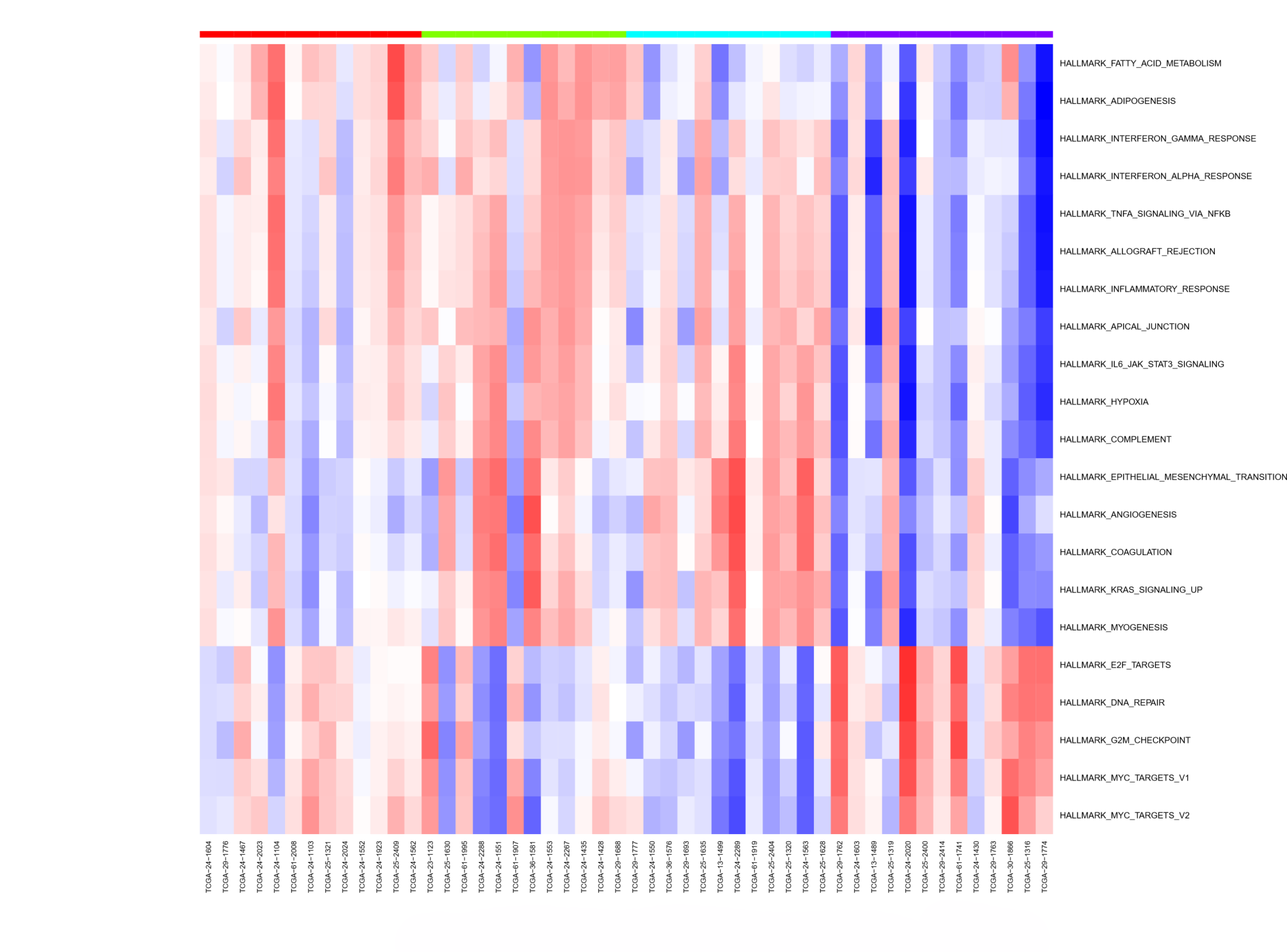
Because our hypothetical example is a classic multi-modal analysis, we used the MFA Analysis and Pathway Analysis workflow developed by NCI and Seven Bridge team. The output from the analysis (see Figure 3) will give you key information on which genetic pathways may be associated with eCRC. (Note: These results are preliminary and intended only as an example to show the output from this workflow.) This entire analysis with a sample size of a few hundred takes less than 1 hour and costs less than $1 to run!
Our example, although hypothetical, shows how you might use the cloud and perhaps overcome some of the barriers standing in the way of using this valuable resource.
Sources
- NCI Cancer Research Data Commons: Cloud-Based Analytic Resources. Cancer Research, 2024.
- Usage of the National Cancer Institute Cancer Research Data Commons by Researchers: A Scoping Review of the Literature. JCO Clinical Cancer Informatics, 2024.
- NCI Cancer Research Data Commons: Lessons Learned and Future State. Cancer Research, 2024.
- Multi-omics Pathways Workflow (MOPAW): An Automated Multi-omics Workflow on the Cancer Genomics Cloud. Cancer Informatics, 2023.
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (40)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (25)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (13)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

Leave a Reply