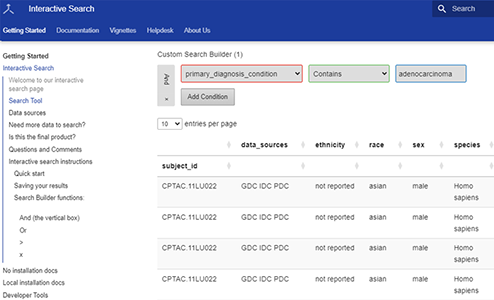
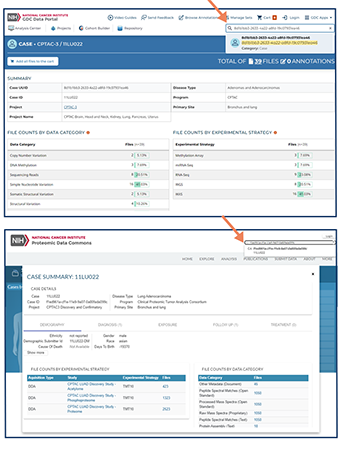
How to Use the Cancer Data Aggregator
What is the Cancer Data Aggregator?
The Cancer Data Aggregator (CDA) is a resource that lets you search for data across NCI’s Cancer Research Data Commons (CRDC). The CDA includes standardized and indexed terms from the Genomic Data Commons (GDC), Imaging Data Commons, Proteomic Data Commons (PDC), Integrated Canine Data Commons, and the General Commons.
This accessible and easy-to-use tool not only allows you to collect data but also explore and analyze that data, making it an invaluable asset if you’re following the cancer data science lifecycle research process. You can find information using harmonized, common language terms. You can then easily work with your search results in Excel, integrate them into a pipeline, or upload to an NCI Cloud Resource.
Say you want to find data for your cancer research. What does that process look like with CDA?
- You have an idea and want to see what data is available.
- You use CDA to search across all data commons.
- You use the IDs to:
- navigate to the data commons hosting the data you’d like to learn more about.
- visit dbGaP and begin the approval process for accessing that data (if any data are controlled access).
- transfer the data to ISB Cancer Genomics Cloud (ISB-CGC), Broad FireCloud powered by Terra, or Seven Bridges Cancer Genomics Cloud (SB-CGC) powered by Velsera to prepare for your analysis.
What Do I Need to Know?
Fundamental Tips for Using the CDA Effectively
The CDA has many features that will help you with your cancer research.
CDA tools for beginners include:
- a point-and-click search tool.
- Google Colab notebooks that provide templates you can use to write queries and run them against the CDA API without the need for installation. This tool is perfect if you’re learning how to code but still need some help.
More advanced CDA tools include:
- a local install of cdapython for a more hands-on experience if you’re comfortable with complex search queries.
And remember, the CDA Team is always available to help! Whether you need assistance with running complex queries, guidance with writing code, general advice, and more, you can reach the CDA Team through the helpdesk or email.
NCI CDA Resources and Initiatives
NCI-Supported Projects
You’ll find CDA contributions in other NCI-supported projects that may be helpful for your cancer research. Explore them to see whether you could benefit from these resources.
- ISB-CGC is an NCI cloud resource that lets you access, explore, and analyze large-scale cancer data through the Google cloud platform. ISB-CGC provides mutation data to CDA, and CDA provides ISB-CGC with aggregated data from across the CRDC. Through ISB-CGC, you can access smaller, easier to use files that ISB-CGC has processed from CRDC data commons. You can also find data through the CDA and then export the data to ISB-CGC to do your analysis. Explore the ISB-CGC BigQuery Table Search to browse tables of metadata and molecular cancer data!
- FireCloud and SB-CGC are NCI-funded cloud platforms you can use for data analysis. Once you’ve found the data you want on CDA, both platforms provide you with user-friendly access to a range of analysis pipelines. Just upload the identifiers you found at CDA to start your analysis.
Publications
If you’d like to learn more about the components of CRDC that make CDA possible, read this American Association for Cancer Research journal article about CRDC core standards and services.
Explore our Cancer Data Science Pulse blog on the CDA if you want to read about the background and development of the CDA.
Additional CDA Resources
- Have specific questions about the CDA? Email the CDA Team!
- Looking for CDA trainings? Bookmark the CDA homepage, which will feature upcoming trainings as soon as they’re scheduled!
- Want to stay up to date on CDA-related news and events? Subscribe to our weekly email updates on cancer data science topics.