Cancer Data Science Pulse
Semantics Series—A Deep Dive Into Common Data Elements
We’re taking a deep dive into Common Data Elements (CDEs)—a key component in CBIIT’s semantic infrastructure (SI). We’ll explore the need for CDEs, the differences between CDEs and terminology concepts for data elements (and how they interrelate), and the advantages of CDEs.
Why do we need CDEs?
CDEs give us a way to standardize and share precise and unambiguous definitions of the meaning of data independent of any data model or data set. CDEs do this in a way that’s structured, predictable, and consistent. Setting and agreeing to a CDE’s meaning and rules before research starts enables us to easily collect, validate, share, and reuse data from different sites, studies, and clinical trials. Unfortunately, it can be nearly impossible to get everyone to agree on the same CDE before beginning data collection. However, even if we set up a CDE after collecting the data, we can still help other researchers understand and reuse those data, providing permanent and detailed semantics moving forward. With a clear CDE roadmap, we can harmonize data across disparate sources, enabling us to conduct further semi-automatic mapping to convert data from one format to another.
Concepts and CDEs—Laying a foundation for meaning
A “concept” describes a unit of meaning—a single idea or unit of knowledge within the context of a specific concept system. Examples of systems are standard terminologies and coding systems (e.g., NCI’s Thesaurus [NCIt], Logical Observation Identifiers Names and Codes [LOINC], and Systematized Nomenclature of Medicine [SNOMED]). Terms in these systems usually have an identifier and a term for referencing the meaning of the concept. Concept terms are not subject to any rules or language limits. Instead, they represent a single idea; for example, “Height,” “Malignant Neoplasm,” or “Love.” We can assemble concepts using linguistic rules to construct sentences and paragraphs that tell (or represent) a specific story.
A CDE refers to a bundle of concepts created within a standard structure to describe the semantics, or meaning, of a data element. At NCI, the structure we use is the International Organization for Standardization’s ISO/IEC 11179 Metadata Registry standard. Specifically designed for managers and owners, it helps them to unambiguously represent the meaning of data.
This registry standard prescribes a set of rules, or grammar, for organizing multiple concepts to fully represent data meaning. For example, the data value “M” is not adequate to convey its meaning, and neither is the concept “Male.” You might wonder, “Male” what? Is it a person’s gender? Spouse’s gender? Canine’s gender? Gender at birth? Gender identity? CDEs help to resolve the question of what was meant by the data “M.” By following the ISO/IEC 11179 grammar/rules for bundling concepts, we can better represent the meaning of data and allow CDEs and concepts to work in concert to tell a story about the data.
A bundle typically includes at least three concepts per CDE. There can be many more if the CDE has an enumerated list of permitted values. Each concept has a specific semantic role.
One concept represents the class of data we’re collecting, such as “Canine” or “Biospecimen.” Another concept represents a characteristic of that class, such as “Gender” or “Weight.” A third concept conveys the category we’re representing, such as “Code,” “Scale,” or “Count.”
An example of using more than three concepts is a CDE that’s restricted to a specific list of permitted values, such as a set of codes or text like “M” and “F.”
In ISO/IEC 11179, each permitted value includes an annotated concept. The CDE information specifies the datatype and other constraints on the CDE, including maximum length or format. In addition, a CDE can define its constraints by pointing to an external standard coding system, such as LOINC or the International Classification of Diseases (ICD). By following the rules of ISO/IEC 11179, we can unambiguously interpret the data value “M,” collected according to a specific CDE, as “The canine’s gender is male,” which is much more expressive than the single concept “Male.”
Persistent semantics over time
When we use a bundle of concepts, each with a specific role, to represent a CDE, it’s much more expressive than using a single concept. Another advantage is that CDEs offer a persistent meaning over time. We must interpret the meaning of a concept within the context of its concept system. Generally, concept systems, and not individual concepts, have versions. New versions of the system can lead to inconsistencies, including dropping or retiring concepts or codes or changing a concept’s name. Some systems could even reuse retired identifiers, resulting in new meanings that are entirely different from the original.
When we use concepts within a CDE, this “drift” in meaning doesn’t happen. From the moment it’s created, each CDE preserves vital information, such as its concept’s preferred name, definition, and identifier.
Because a CDE has its own persistent unique identifier, including version, you can trace the semantics of the data over time. This serves as a snapshot of the concept, and thus the CDE’s meaning, for that specific version. Therefore, unlike a concept in other systems, the CDE’s meaning cannot shift with the release of a new version of the concept system. The data’s definition remains the same, independent of changes to concept systems or even data models.
Mapping using CDEs
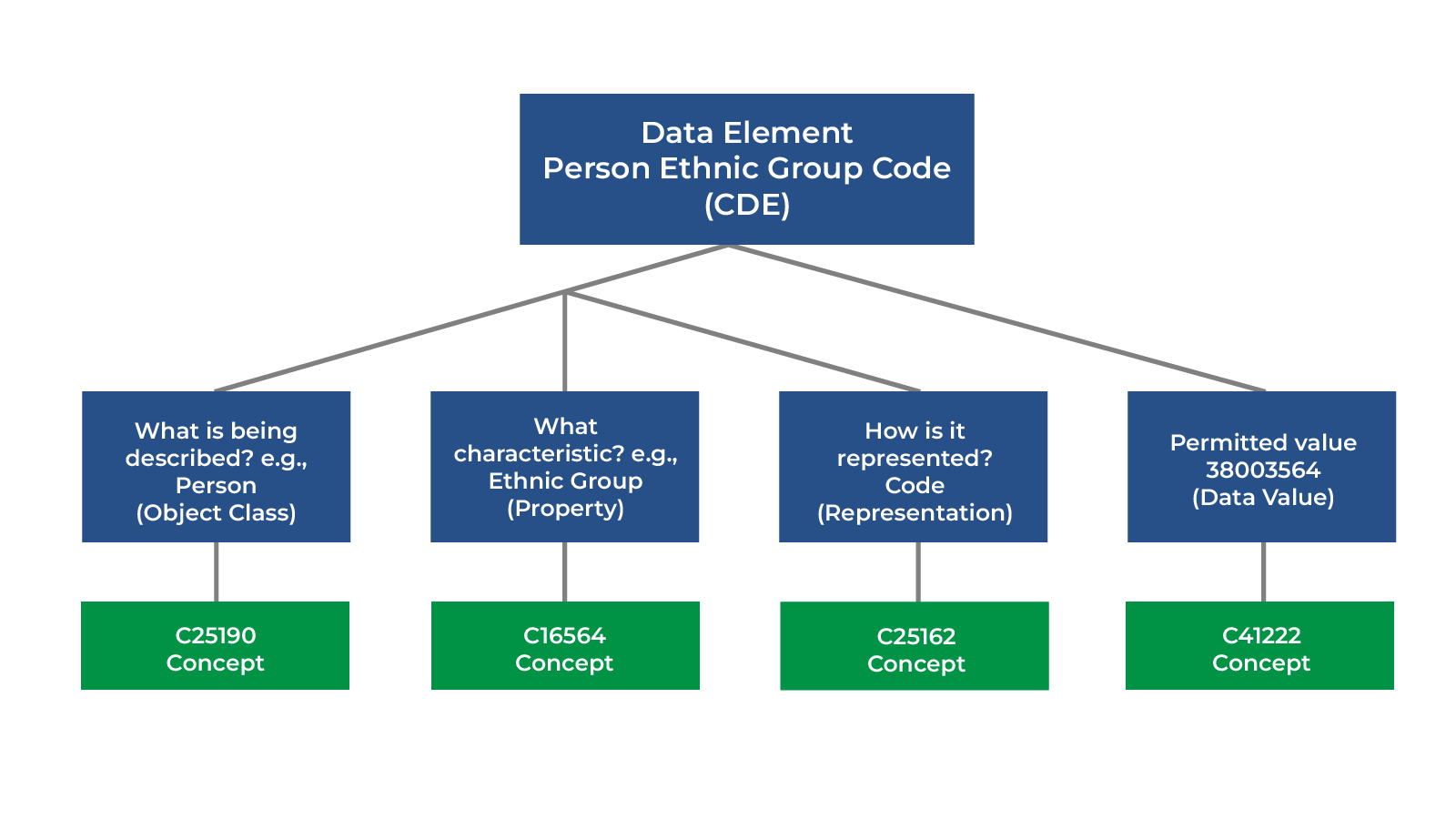
Using CDE concept information, we can map across different CDEs that rely on different standard terminologies or coding systems. An example of this mapping is the way disparate data models capture a person’s ethnicity with different coding systems. It can be very time consuming (or even impossible) to research, map, and transform data, especially in cases with proprietary or local coding systems. The example below shows how concepts can help harmonize data across four disparate data models, capturing a person’s ethnicity using different coding systems:
- OMOP (Observational Medical Outcomes Partnership) data value using its own concept identifier “38003564”
- FHIR (Fast Healthcare Interoperability Resources) data value using the HL7 Value Set code “2186-5”
- CDISC (Clinical Data Interchange Standards Consortium) data value using a text value of “NOT HISPANIC OR LATINO”
- PCORnet (Patient-Centered Clinical Research Network) data value using the indicator “N”
Using a CDE defined for each data model and annotating the meaning with “C25190” (meaning “Person”), “C16564” (meaning “Ethnic Group”), and the permitted values within the NCIt concept codes, we can determine that the data value is associated with “C41222,” which means “Not Hispanic or Latino.”
This information allows humans and computers alike to determine that 1) the four data elements pertain to the same thing—a person’s ethnic group, and 2) the data values have the same meaning—“Not Hispanic or Latino.” This example illustrates how a CDE’s bundle of concept annotations (in a pre-defined structure) supports machine and human semantic interoperability and data transformation, allowing us to map across disparate data models.
New technologies and the future of CDEs
The CBIIT SI team offers new tools and services to help researchers find, create, share, and reuse CDEs aimed at simplifying and expanding the use of these rich semantic bundles across research and healthcare.
For example, we’re working now to establish a common semantic core to facilitate harmonization across national data models, U.S. Food and Drug Administration reporting requirements, and healthcare. Through these initiatives, the aim is to make more data available for aggregation and reuse in research.
In addition, with the development of artificial intelligence, and advances in ontology technology, we’re looking to leverage the semantics offered by CDEs to help expand our knowledge from research and healthcare findings.
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (40)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (25)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (13)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

Leave a Reply