Cancer Data Science Pulse
Semantics Primer
NCI’s Center for Biomedical Informatics and Information Technology (CBIIT) has long been concerned with building interoperable systems to support NCI’s mission of leading, conducting, and supporting cancer research. Semantic interoperability is an important component of these systems and is vital for integrating and retrieving data—whether those data are being accessed by humans or through an Application Programming Interface (API).
In the broadest sense, semantics allow us to “speak the same language.” In science, consistent use of terminology with an understood shared meaning (the semantics) gives us a foundation for ensuring that researchers within a lab or across the world can communicate and accurately compare and reproduce critical findings.
This is particularly crucial for the use and re-use of data found in community resources, such as NCI’s Cancer Research Data Commons, which includes a wide variety of data—from studies on genomics and proteomics to imaging and clinical information.
Basic Concepts
Words such as “semantics,” “concept,” and “ontology“ are often used when developing terminologies, such as the NCI Thesaurus (NCIt) for cancer-related terms. The term “semantics” typically is defined as the study of meaning as applied to a particular context or field (such as language).
For our purpose, we use the term semantics to denote meaning, or, in association with other terms, to describe knowledge-based systems or resources.
The term “concept” is used to denote a unit of meaning. The definition that we have in the NCIt is “a discrete notion having a single meaning.” Alternative definitions can be found in international standards for terminology development, for instance, unit of knowledge and unit of thought.1
When building a concept-based terminological system, such as the NCIt or the Unified Medical Language System (UMLS) Metathesaurus, a concept typically is represented by a variety of both simple and compound terms and synonyms that denote the same meaning.
As an example, Figure 1 shows details of a concept in the NCIt, identified by the code C17387. This one code yields a variety of terms that are synonymous with “cellular tumor antigen p53,” including “antigen NY-CO-13,” “tumor suppressor p53,” “tumor protein p53, “p53 protein,” and simply “p53.”
Of note, in this case, some individual terms (“tumor,” “p53,” and “protein”) occur more than once. But these common terms are not shared across the board by all the synonyms. This is not always the case; for instance, with the concept C7414 (“bladder villous adenoma”), “adenoma” is found in all the synonyms. The example of p53 illustrates that concepts often include a variety of terms of interest, all of which need to be considered when constructing a vocabulary.
Because it reduces ambiguity, capturing all the terms that reflect a single concept facilitates integrating data from different sources. This allows researchers to perform secondary analyses and to re-use data well beyond their original investigations.
The term “ontology” describes something more complicated than a terminology. NClt defines ontology in information science as “an explicit formal specification of how to represent the objects, concepts, and other entities that are assumed to exist in some area of interest and the relationships among them.”2
Most important in terms of technology, this formal declaration of concepts and their meanings (such as provided by Description Logic) makes their definitions machine interpretable. Figure 2 shows an example of a logical definition for the concept C9003, “adrenal cortex adenoma,” in the NCIt.
The formalism (that is, the notation, and its structure, in which information is expressed) used in this logical definition is the OWL Description Logic (the Web Ontology Language).
Pay for What You Need
Collecting and assigning terms in a field of study or domain is only one aspect of what’s required to build a useful ontology or terminological system.
Depending on how the system is used, a simple list of terms might be enough, such as when describing a common data set or retrieving data that all share a similar characteristic. In those cases, the terms might not need textual definitions, because everyone using this system would already be familiar with the concepts and agree on how they are being used. Or perhaps, a data set is very narrow and, thus, needs only limited vocabulary.
In other cases, however, a complex ontology or terminological system is necessary. Such a system would be needed when a variety of characteristics require annotations or when there may be a need for downstream integration. With such complex systems, we can’t simply add more terms and hope that this will cover every possible need. Other features come into play, such as synonyms, abbreviations, textual definitions, and (if necessary) machine-interpretable definitions, which ensure the terms or concepts are clear. They can facilitate mapping to entries in other vocabularies and to annotations in repository data.
Seeking a Vocabulary System for Data Science
Not surprisingly, the complexity of data science requires a comprehensive vocabulary system that can grow and evolve as the field changes.
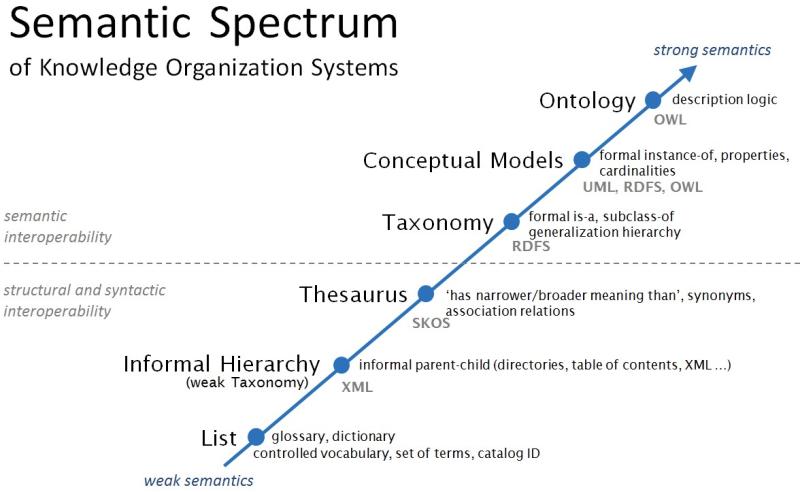
Not every vocabulary and terminology is suitable for this purpose. Terminological systems suitable for data science fall in the more expressive, semantically strong, and higher interoperability side of the semantic spectrum (see Figure 3). Such terminologies are structured, at the very least, in taxonomies with a formal hierarchy; whereas the semantically strongest, that is ontologies, utilize logic-based formalisms to define the concepts.
But for data science especially, interoperability is vital. In fact, three of the four elements needed to meet the basic tenets of FAIR data (data that are Findable, Accessible, Interoperable, and Reusable) depend on semantically rich metadata and terminologies. Ontologies or semantically strong terminological systems are essential.
Our goal in using semantics for data science is to develop systems that will allow researchers to query, retrieve, and combine very different data sets and still allow for extensive analysis.
No doubt, as data science continues to grow, the role of semantics also will evolve. The structures we put into place now will need to be as dynamic as the field to keep up with these rapid advances.
This primer offers just a snapshot of how semantics are currently structured for use in data science. Future blogs will examine the importance of building a consensus when tackling semantics and give greater detail on the infrastructure and tools available from NIH/NCI/CBIIT.
1 The existence of different definitions for "concept" has been used as a criticism of concept-based ontologies.
2 This definition is slightly modified from Gruber, “Ontology,” Encyclopedia of Database Systems, Ling Liu and M. Tamer Özsu (Eds.), Springer-Verlag, 2008, pp 1963-1965.
Leave a Reply
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (40)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (25)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (13)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)



Deb Hope on January 14, 2022 at 02:58 p.m.