Cancer Data Science Pulse
Proteomic Data Commons (PDC)—A Resource for Probing Today’s Cancer Questions
Genes carry the body’s blueprint and, as such, set the stage for building every cell, tissue, organ, and organ system. But it’s the body’s proteins that extend this blueprint into action. This is especially evident with cancer, where proteins direct tumor growth, including how invasive it is, whether it spreads, how it interacts with cells around it, and how it responds to treatment.
Proteomics data offer a snapshot of expressed proteins at a given time and within a given individual. These data also help to overcome one of the chief limitations of the genome by giving us a way to tease apart the uniqueness of each cancer case. This is the basis behind precision medicine—treating each person’s cancer in a highly targeted way. Amassing data from multiple labs in one place will enable us to better understand not only how genes translate to the proteins but how those proteins are linked to disease and health.
The next step in the field of proteomics is to make the proteomic research data that are accumulating from labs across the nation and the world available in one place and to make that data fully accessible to a broad range of users.
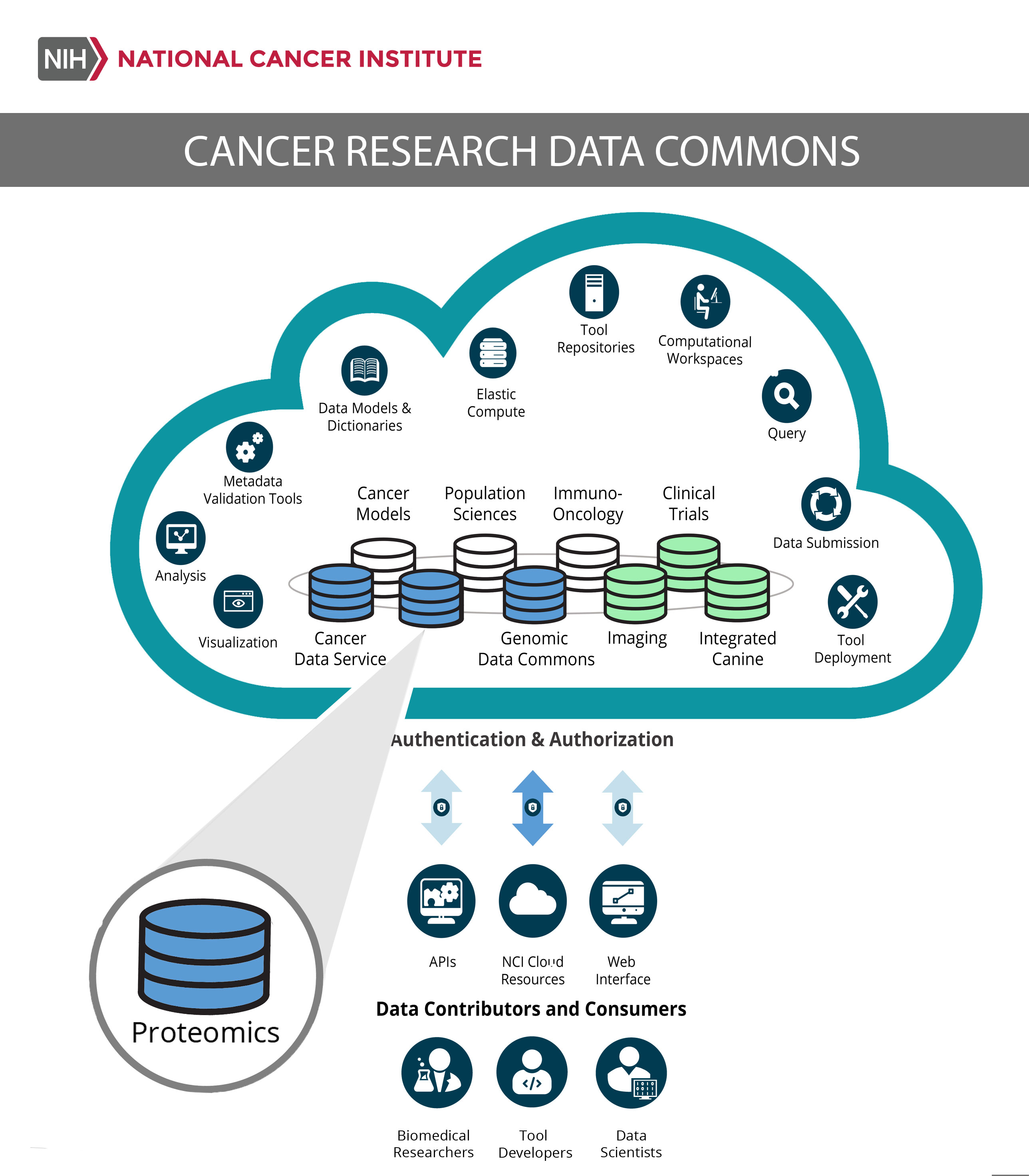
This is the goal of NCI’s Cancer Research Data Commons (CRDC), a cloud-based resource designed by NCI’s Center for Biomedical Informatics and Information Technology (CBIIT) to link data with people—both to speed cancer research and to advance treatment through the use of precision medicine.
The Proteomic Data Commons (PDC) gives researchers and the public access to three types of proteomic data: mass spectra, identified peptides, and protein reports, as well as clinical, biospecimen, and other metadata (such as sample type, tumor grade, diagnosis, and patient demographics). Moreover, all proteomic data are open access and can be freely downloaded and shared.
The database currently houses 64,720,831 spectra, 991,578 peptides, and 12,704 proteins and it’s still growing. The foundation of this proteomic information came from samples collected as part of the NCI’s Clinical Proteomic Tumor Analysis Consortium (CPTAC). That network includes information on tumors from colorectal, ovarian, clear cell renal cell carcinoma, uterine corpus endometrial carcinoma, lung carcinoma, and breast tumors.
As the PDC matures, additional cancer proteomic datasets will be added, including data from other large proteomic programs, such as the Children's Brain Tumor Tissue Consortium (CBTTC) project and the International Cancer Proteogenomic Consortium (ICPC).
A unique feature is PDC’s connection to other NCI Cancer Research Data Commons nodes, such as the Genomic Data Commons (available now) and Imaging Data Commons (under development). This vast data network aims to aggregate and harmonize datasets to produce even more robust results.
Using the PDC
The PDC is now available for queries and analysis of publicly accessible datasets. Data can be searched at a variety of levels—by program; organ site; analytical fraction; or by molecular features, such as a gene or protein. Browsing is supported by a web interface for humans and through an application programming interface (API) for machines.
In addition, PepQuery has recently been added to the PDC website to enable users to customize their spectra searches. It allows for quick and easy proteomic validation of genomic alterations through a series of five drop-down menus. Using PepQuery, investigators can identify novel peptides and their splice junctions, as well as single amino acid variants.
A tutorial on navigating the database is available from Seven Bridges, one of NCI’s Cloud Resources. Using the Seven Bridges “Data Cruncher,” and a specially designed PDC metadata file (PDC_metadata.json) that’s available in the public reference resources, users can extract data from the PDC. Other NCI Cloud Resources include the Broad Institute’s FireCloud and the Institute for Systems Biology’s Cancer Genomics Cloud.
A special Proteomic Data Commons Webinar will be held on November 13, 2019, from 1:00 p.m. to 2:00 p.m. ET. Registrants are invited to submit questions prior to the webinar. Please send questions to LaToya Kelly with the subject line OCCPR Webinar: PDC. Questions should be submitted by noon on Tuesday, November 12, 2019, and will be addressed on a first-come basis, time permitting.
We also invite you to explore the available datasets, submit proteomic data, and make suggestions for new software tools. Please send feedback to the PDC Helpdesk.
Categories
- Data Sharing (64)
- Training (39)
- Informatics Tools (39)
- Data Standards (35)
- Genomics (35)
- Precision Medicine (32)
- Data Commons (32)
- Data Sets (26)
- Machine Learning (24)
- Seminar Series (22)
- Artificial Intelligence (21)
- Leadership Updates (13)
- Imaging (12)
- High-Performance Computing (HPC) (9)
- Policy (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Request for Information (2)
- Information Technology (2)
- Awards & Recognition (2)
- Publications (2)
- Childhood Cancer Data Initiative (1)

Leave a Reply