Cancer Data Science Pulse
Cancer Data Harmonization—Setting Standards and Breaking Down Silos
The biomedical sciences have changed a lot over the past decade as the data have evolved to be ever larger, more distributed, more interdependent, and more natively web-based. This transformation has fundamentally diversified both the actors doing the analyses and the types of data accessed.
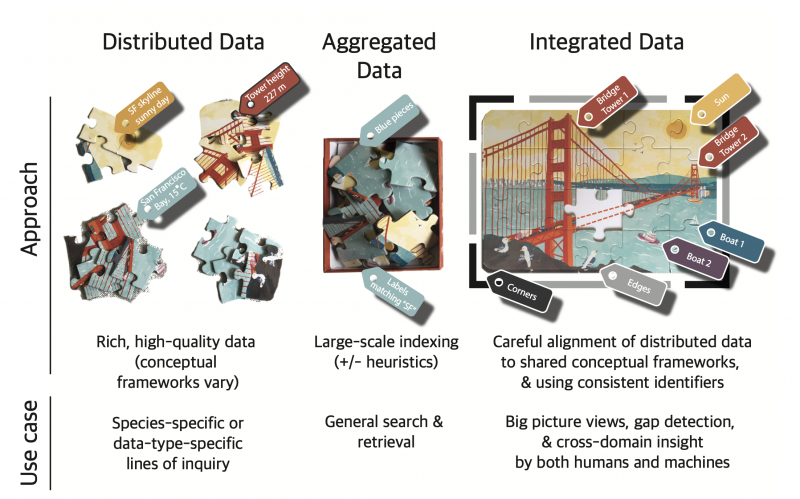
To answer more complex scientific questions by a greater breadth of stakeholders, we need standards and systems that are up to this challenge. Pooling data from numerous sources strengthens the power of the information, but only if it can be meaningfully connected. Merely aggregating the data is not generally useful in itself. A firm foundation is needed so the petabytes of data generated from cancer research can be combined, analyzed, and turned into knowledge that’s useful to the broader research community.
Harmonization starts with careful organization. This can be as simple as standardizing names so that all the sources of data use the same terms. Or it might involve setting up data “fields” so that each piece of information in a new data source can be easily merged with the existing data.
The level of difficulty in harmonizing data grows exponentially when we aim to consolidate information from very different sources, such as that stored in NCI’s Cancer Research Data Commons (CRDC). These rich storehouses of data give researchers, clinicians, and others the opportunity to blend findings as diverse as histology and genetics, and to match those findings with data from epidemiology and demographics.
Imagine you want to run an analysis to determine the number of post-menopausal women who have HER2-positive breast cancer to see how they responded to a particular medication. Such a query would need to be able to reach into databases that store genomic, proteomic, tissue, and demographic data and extract data used to generate meaningful results.
A new initiative, the Center for Cancer Data Harmonization (CCDH), is NCI’s next step in helping to link these datasets. The CCDH will draw on established data standards as well as feedback from the research community to develop new ways of organizing data. Other tools to support harmonization also are being developed by the CCDH, including a special concierge-type service to help CRDC nodes and NCI-supported Data Coordinating Centers in submitting and organizing data and a web-based portal for easy access.
How are Data Harmonized?
Today’s data landscape has repositories in different places, containing a wide variety of information. These “data communities” often have been built around key initiatives such as those collecting information on genes, proteins, demographics, and clinical characteristics.
Key Steps include:
- Standardize the names. This ensures that everyone refers to certain concepts or data in the exact same way. To do this, NCI draws on a number of existing data standards, including the NCI Thesaurus terminology, the NCI Metathesaurus, CDISC Terminology, DICOM, among others.
- Assign durable identifiers.1 Like a fingerprint, an identifier (often called a globally unique identifier or GUID) allows users to uniquely identify data items from a variety of sources and then combine them for stronger analysis.
- Curate data.2 Annotate data to meet specific standards, such as annotation to ontologies or adherence to specific data models.
- Harmonize data. To be able to use data en masse, it is necessary to harmonize the data to make individual datasets more than the sum of the parts. However, this is laborious, as different sources use different models or schemas. Battles must be picked based on what is the best fit for purpose. Resources such as Fast Healthcare Interoperability Resources (FHIR), Clinical Data Interchange Standards Consortium (CDISC)/BRIDG, and Biolink-Model can provide a backbone for harmonizing fields, common data elements, and value sets.
- Test and test again. Here data are further filtered to remove information that is out of step, such as outliers or data that were entered incorrectly or processed in a sub-standard way. Testing also evaluates the reproducibility of the data.
- Share, attribute, and document. Making the data accessible within the individual commons as well as across the different Data Commons ensures full availability for analysis. Provenance of where the information came from, when, and who touched it is critical for using data in aggregate. It is also important to provide attribution, potentially using new systems such as the Contributor Attribution Model.
Future Directions
As the CRDC grows and as researchers seek to add and analyze new and larger data sets, standards will help bring data together in a more consistent way for improved analysis. And just as research informs practice, findings from the frontlines of patient care also need to feed back into the system to drive new research questions. Joining with the cancer research community, we can realize the dream of a harmonious cancer data ecosystem.
Learn more about the CRDC and the data being made accessible by the CCDH.
Key Objectives for the CCDH:
- Create a harmonized data model and provide cross-mapping to domain-specific node data models.
- Provide semantic concierge services to CRDC nodes and NCI Data Coordinating Centers related to data models, metadata, and terminology.
- Implement a web-based portal.
- Create, adapt, and disseminate data harmonization tools for use by CRDC nodes and NCI Data Coordinating Centers.
- As needed, develop new terminology, metadata, mappings, and models to support data aggregation through the CRDC.
Special thanks to Mr. Bron Kisler, former Biomedical Informatics Specialist with the National Cancer Institute’s Center for Biomedical Informatics and Information Technology, for his assistance in the preparation of this blog.
1 McMurry, J.A., Juty, N., Bloomberg, N., Burdett, T., Conlin, T., Conte, N., et al. Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data. PLoS Biol. 2017;15: e2001414.
2 Tang, Y.A., Pichler, K., Füllgrabe, A., Lomax, J., Malone, J., Munoz-Torres, M.C., et al. Ten quick tips for biocuration. PLoS Comput Biol. 2019;15: e1006906.
Categories
- Data Sharing (65)
- Informatics Tools (41)
- Training (39)
- Genomics (36)
- Data Standards (35)
- Precision Medicine (34)
- Data Commons (33)
- Data Sets (26)
- Machine Learning (24)
- Artificial Intelligence (23)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (12)
- Policy (9)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Information Technology (2)
- Childhood Cancer Data Initiative (1)

Leave a Reply