Cancer Data Science Pulse
Informatics Technology for Cancer Research Program Drives and Fosters Community of Cancer Informatics Researchers: An *AMARETTO Tool Success Story
Researchers in cancer informatics are challenged by a profusion of data resources, literature, and computational tools. Mechanisms for breaking down silos, increasing communication, and fostering collaboration are difficult to build and sustain, even in individual institutions. Owing to its impact on my own research on *AMARETTO, I view the Informatics Technology for Cancer Research (ITCR) Program as a major step forward in successfully solving these challenges.
ITCR is a trans-NCI program that supports investigator-initiated, research-driven informatics technology development spanning all aspects of cancer research. Unlike typical research approaches, ITCR places an emphasis on engaging users and investigators in the cancer research community in a variety of methods, including:
- Monthly conference calls and annual meetings of investigators working in a variety of domains spanning tumor genetics, genomics, and imaging, which has exposed me to projects and their initiators in ways that don't arise in typical approaches to research.
- Funding opportunities foster various stages of the informatics technology development lifecycle, including algorithm development, prototyping and hardening, enhancement and dissemination, and sustainment, with administrative effort specifically directed at identifying interconnections between independent projects and set-aside programs to fund specific collaborative proposals.
- Monthly ITCR Working Groups focus on training and outreach, technical, and sustainability and industrial partnership aspects.
- Webinars, demos, and workshops of its informatics technologies at cancer research conferences.
To inspire the community of cancer informatics researchers, here I highlight how the ITCR Program has successfully impacted the development, dissemination, and general applicability of *AMARETTO, and has led to broadly catalyzing and accelerating new discoveries in cancer.
The Development of *AMARETTO
The *AMARETTO framework provides software tools for network biology and medicine, towards a data-driven platform for diagnostic, prognostic, and therapeutic decision-making in cancer. Specifically, *AMARETTO offers modular and complementary solutions to multimodal and multiscale aspects of network graph-based fusion of multi-omics, clinical, imaging, and driver and drug perturbation data across studies of patients, etiologies and model systems of cancer.
The ITCR Program supports the development of *AMARETTO to identify novel mechanisms of viral carcinogenesis and uncover new therapeutic targets for chemoprevention of hepatocellular carcinoma. Through a collaborative set-aside with Vincent Carey, ITCR also supports Bioconductor-embedded developments of *AMARETTO as an imaging genomics tool for diagnostics and therapeutics in hepatocellular carcinoma and glioblastoma multiforme.
The conceptualization of *AMARETTO is co-led by Olivier Gevaert, Mikel Hernaez and myself, and has grown towards a multidisciplinary investigational team of cancer informaticians, biologists, and clinicians, ultimately leading to a continuously expanding network of informatics technology-driven collaborative initiatives to accelerate biomedical research and healthcare delivery, for better diagnosis and therapy of human disease. Originally formulated for studies of cancer, we are reformulating these tools for applications to other complex diseases; for example, for neurological and immune-mediated diseases in collaboration with my colleagues Jishu Xu, Nikolaos Patsopoulos, Anna Krichevsky, Erik Uhlmann, Francisco Quintana, Vijay Kuchroo and Howard Weiner.
*AMARETTO Supports New Cancer Discoveries
To demonstrate its utility, working with my collaborator Thomas Baumert, we leveraged *AMARETTO to discover novel therapeutic treatments for chemoprevention of hepatitis C and B virus-induced hepatocellular carcinoma. Several predicted drug compounds were successfully validated in vivo as demonstrated by their ability to inhibit liver fibrosis and inflammation with prevention of cancer development in animal models. Thus, they potentially represent a safe and low-cost approach for chemoprevention of hepatocellular carcinoma across viral and other etiologies. For these types of cancer informatics applications, the ITCR Program has recently launched the Working Group on Sustainability and Industrial Partnership to help guide its investigators in successful transitioning of informatics technologies to applications in clinical practice and within industrial partnerships. A startup Alentis Therapeutics has been created to develop novel molecules for treatment of advanced liver disease and cancer.
ITCR Catalyzes the Dissemination and Hardening of *AMARETTO
The ITCR Program already identified and sustained GenePattern's team led by Jill Mesirov and Michael Reich to support Notebook-oriented interfaces for dissemination, and Bioconductor’s team led by Vincent Carey, Martin Morgan, and Levi Waldron as a vehicle for dealing with testing, performance evaluation, and continuous integration of the underlying code. These ITCR-initiated collaborations have catalyzed the dissemination of *AMARETTO as user-friendly tools via GenePattern and Bioconductor, and GenePattern and Jupyter Notebooks for several case studies of cancer, including virus-induced hepatocellular carcinoma, glioma and glioblastoma, and pan-cancer studies (EBioMedicine 2018).
Future *AMARETTO Impact Through ITCR
To broaden its impact, our team continues to establish connections to other informatics tools supported by ITCR, including interrogating novel genetic and epigenetic heterogeneity discovered from cancer transcriptomes assembled from individuals or single cells by Trinity CTAT (led by Aviv Regev and Brian Haas) in *AMARETTO networks, disseminating networks via the interactive network database NDEx (led by Trey Ideker and Dexter Pratt) and dynamic interactive heatmaps for interpretation of networks with NG-CHM (led by Bradley Broom). In parallel, the *AMARETTO team will continue to follow the ITCR’s guidelines for transitioning *AMARETTO to applications in clinical practice and within industrial partnerships, such as applications of drug discovery focused on validation through clinical trial studies.
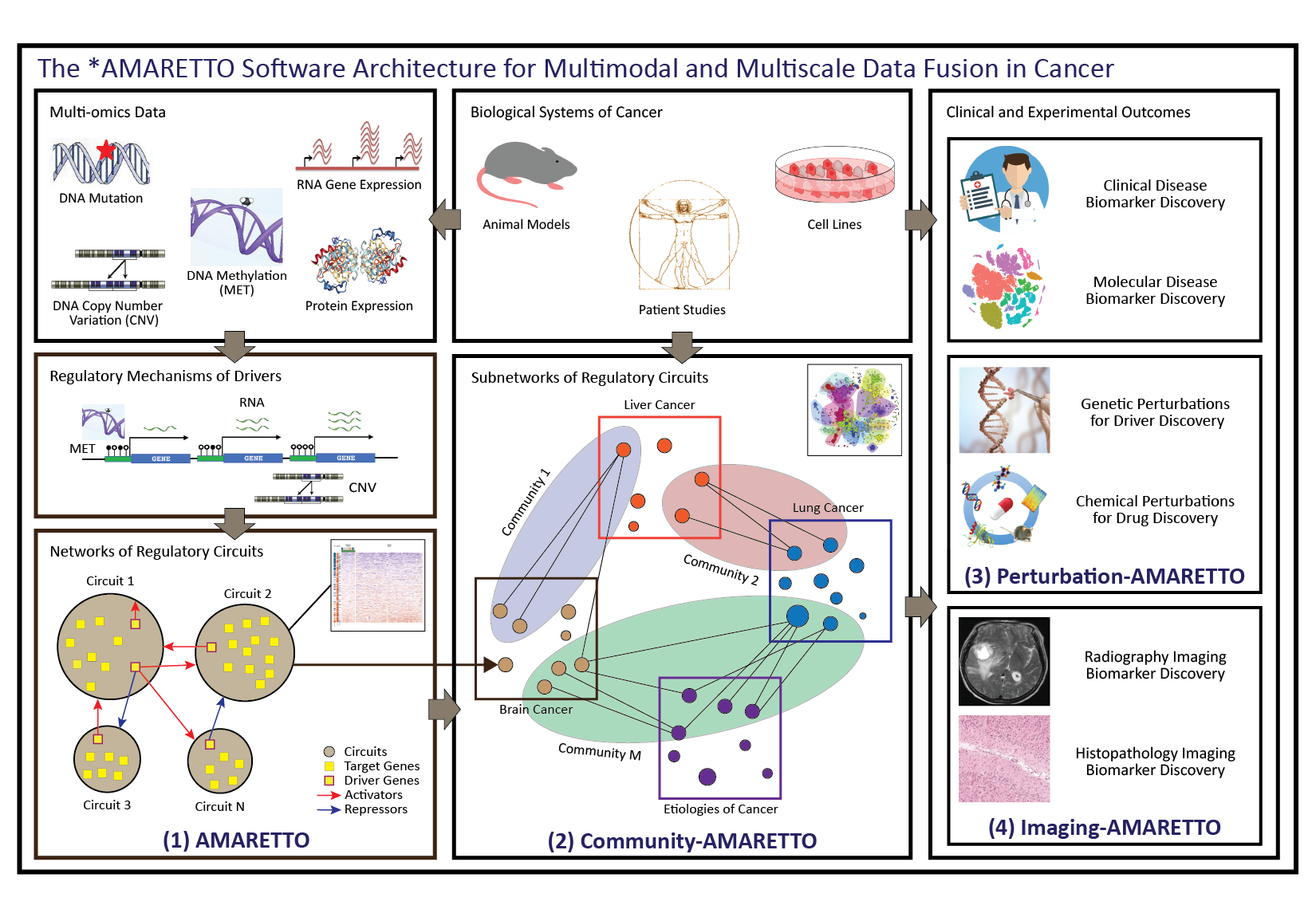
The *AMARETTO software architecture. The *AMARETTO framework provides software tools for network biology and medicine, towards a data-driven platform for diagnostic, prognostic and therapeutic decision-making in cancer. The *AMARETTO platform offers modular and complementary solutions to multimodal and multiscale aspects of network graph-based fusion of multi-omics, clinical, imaging, and driver and drug perturbation data across studies of patients, etiologies and model systems of cancer. Specifically:
(1) The AMARETTO algorithm learns networks of regulatory circuits - circuits of drivers and target genes - from functional genomics or multi-omics data and associates these circuits to clinical, molecular and imaging-derived phenotypes within each biological system (e.g., model systems or patients);
(2) The Community-AMARETTO algorithm learns subnetworks of regulatory circuits that are shared or distinct across networks derived from multiple biological systems (e.g., model systems and patients, cohorts and individuals, diseases and etiologies, in vitro and in vivo systems);
(3) The Perturbation-AMARETTO algorithm maps genetic and chemical perturbations in model systems onto patient-derived networks for driver and drug discovery, respectively, and prioritizes lead drivers, targets and drugs for follow-up with experimental validation; and
(4) The Imaging-AMARETTO algorithm maps radiography and histopathology imaging data onto the patient-derived multi-omics networks for non-invasive radiography and histopathology imaging diagnostics.
Credits to *AMARETTO team members: Mohsen Nabian, Artur Manukyan, Celine Everaert, Shaimaa Bakr and Jayendra Shinde.
Categories
- Data Sharing (64)
- Training (39)
- Informatics Tools (38)
- Genomics (35)
- Data Standards (34)
- Data Commons (32)
- Precision Medicine (31)
- Data Sets (25)
- Machine Learning (23)
- Seminar Series (22)
- Artificial Intelligence (20)
- Leadership Updates (13)
- Imaging (12)
- High-Performance Computing (HPC) (9)
- Policy (9)
- Jobs & Fellowships (7)
- Funding (6)
- Proteomics (5)
- Semantics (5)
- Information Technology (2)
- Awards & Recognition (2)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

Leave a Reply