Cancer Data Science Pulse
It Takes a Village: The Value of Collaboration in Genomic Cancer Studies
As scientists, we know that scientific discovery doesn't take place in a vacuum, although there are times late at night in the lab or behind a laptop that it certainly may feel that way. Ideas can be sparked anywhere and anytime—at a professional conference, during a commute, even during a long run after work. But to truly bring those ideas to fruition, and to advance a hypothesis to the next level, we need to collaborate.
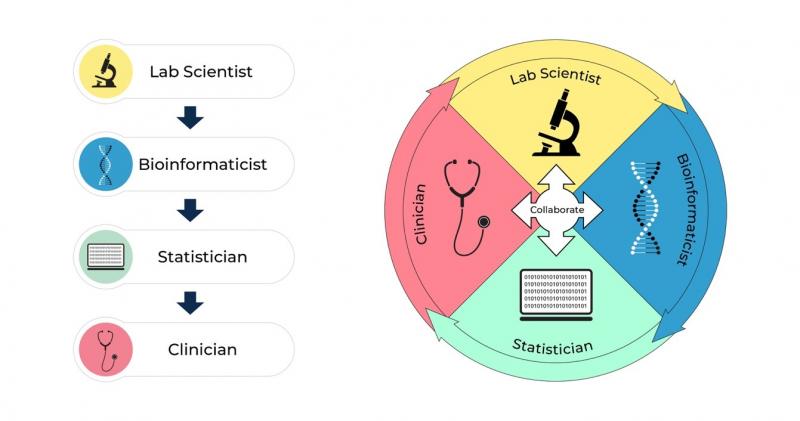
The need for collaboration is especially apparent in the field of genomic medicine. The explosion of genetic information and direct access to large-scale genomic data not only opens up new areas for exploring today's most pressing research questions, it also serves as a reminder of the importance of collaboration at every stage of the study—from obtaining samples from patients and producing data, to performing bioinformatic and statistical analysis, and, ultimately, to clinical interpretation. We need to involve the full team of specialists along this work pipeline or risk negative effects on the study’s outcome.
Such collaborations are not without obstacles, though. Identifying experts in these fields, engaging them in the study, educating them on the nuances of the science and technology, ensuring proper recognition for all team members, working onsite and remotely, and breaking down barriers related to schedules, priorities, and funding—all can derail even the best-conceived project.
Limited collaboration often leads to experts who perform their specialized work in isolation (the so-called "siloed" approach). And it can be detrimental. When inadequate communication exists between experts in the various domains, this can result in questionable or inaccurate interpretations of the data. This is especially true when the data are intended for clinical application, potentially creating distrust among those we most hope to help.
The successful translation of findings from the bench to the bedside hinges on our ability to communicate. Many clinicians are eager to better understand how to apply clinical genomic findings and may be uncertain as to how to apply genomic knowledge to patient care. Healthcare providers not trained as geneticists, who may be wary of the data to begin with, may be reluctant to trust these findings, impacting those on the frontlines of patient care.
Scientists in the genomics field have successfully collaborated on several key projects. Highly successful interdisciplinary collaborations have occurred all along the genomics pipeline—from obtaining specimens from patients to clinical application—in large scale, well-managed projects such as The Cancer Genome Atlas (TCGA). This massive collaborative effort, guided by the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), brought together researchers with diverse expertise from multiple institutions. The outcome has been the characterization of more than 20,000 primary cancers with matched normal samples for 33 types of cancer. Thanks to this effort, we now have well-curated data sets that are available to a wide community of researchers.
From the start, the person designing the clinical and laboratory components of the study needs to be mindful that different sets of algorithms might be applied at different stages of the bioinformatic pipeline. Additionally, the bioinformatics experts working on the study need to be aware of how the data were generated in the laboratory. Upon completion of the bioinformatics analysis, several statistical methods can be applied to correct the raw analytic findings, minimizing the chances of false-positive results. A key concern with such large data sets is the challenge of multiple testing. Non-statisticians need to understand that statistical analysis in the context of multiple testing can threaten the results of the study, leading to false-positive interpretations. A statistician should be involved in the development of the protocol and have a chance to suggest the best approach, from the initial study design to downstream analyses.
One way to keep everyone "in the loop" is to apply uniform standards that provide direction on how the study will be developed and how the findings will be interpreted. A given pipeline should have its own allotted algorithms, and these should be documented in standard operating procedures. Enforcing these standards will ensure that full consensus exists across the research team with regard to how the genomics data will be interpreted—from the initial concept and throughout the study. Such guidelines also will help the bioinformatics team to better characterize a disease (risk, interventions, and opportunities for targeting treatment through precision medicine).
Also, rather than thinking of the genomic pipeline in a traditional sense, flowing linearly from its source to a final endpoint, we propose a path that's circular, or iterative, and allows for input and feedback from all team members at every stage of the study (see figure). Using this model, principal investigators can design studies that yield meaningful results. This enables them to set clear goals and to establish methodology before the study begins. Such a prospectively planned approach is infinitely preferable to retrospectively implementing the analytic approach after the upstream processes have been enacted and completed.
Summary
Applying genomics research in clinical decision-making hinges on careful collaboration among experts specializing in multiple disciplines. From specimen collection and data production in the laboratory through bioinformatic analysis and statistical evaluation—it takes a team approach. Most importantly, the data require judicious interpretation. Dynamic and ongoing interactions within the team will enable us to keep the "end goal" in mind and to produce results that are most meaningful to the clinicians and patients we hope to serve.
Source material for this blog originally appeared in the "Value of collaboration among multidomain experts in analysis of high-throughput genomics data," by D. Meerzaman and B.K. Dunn, Cancer Research; American Association of Cancer Research, 2019.
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (40)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (24)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (12)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

Leave a Reply