Cancer Data Science Pulse
Wrangling Data for Microbiome Research—Focus on QIIME 2
Today, we’re experiencing a boom in microbiota data generation. Finding new ways to manage, integrate, and analyze that data is vital to expand our understanding of the links between the human microbiome and cancer, a research area that is rapidly gaining in importance. Two DNA sequencing techniques have dominated microbiome research to date:
- In marker gene (or amplicon sequence) studies, a specific genomic region common to all microbial organisms is used as a single “genetic fingerprint” to identify the organisms present in a sample. These studies have been the workhorses of microbiome research. One gene in particular, the 16S ribosomal RNA (or simply 16S), has been used to generate taxonomic profiles across a broad cross-section of microbial communities. This method alone has given us the ability to identify far more organisms than the traditional method of culturing. The 16S gene not only has helped us understand the human microbiome, but it has given us insight into the microbial world as a whole and shown that it’s far more diverse than previously thought.
- In shotgun metagenomic studies, rather than targeting a single gene (like the 16S gene), all DNA extracted from a sample is sequenced. Shotgun sequencing gives us more detail than a marker gene survey, because it can provide the full genome sequences of the organisms present in a sample. Moreover, this type of sequencing can reveal the functional potential of a microbiome, so we not only can see “who” is there but also what they might be doing. However, shotgun metagenomic sequencing is currently limited by several factors, including higher cost and more computationally intensive data analysis workflows relative to marker gene sequencing.
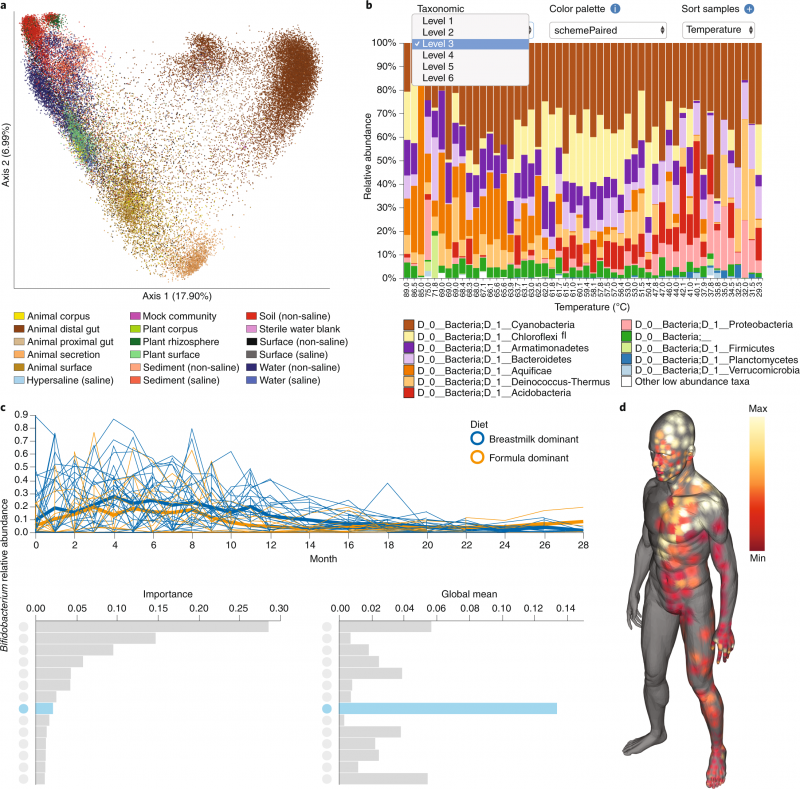
Deriving new knowledge from these techniques has been an ongoing challenge for researchers and data scientists alike. The QIIME microbiome bioinformatics platform, collectively referring to QIIME 1 and 2, was initially developed to address that need for marker gene data. The platform takes users from raw sequencing data through interactive visualizations and publication-quality results. (See Figure for examples of visualization tools.) With support from NCI’s Informatics Technology for Cancer Research (ITCR) program, QIIME 2 is now expanding to support shotgun metagenomics data analysis, as well as other “microbial-omics” data types. QIIME 2, which officially succeeded QIIME 1 in January 2018, was initially funded by the National Science Foundation, followed by the Chan-Zuckerberg Initiative, and most recently by ITCR.
Since its inception, QIIME has become central to microbiome research, amassing nearly 30,000 citations in diverse scholarly journals over the past decade (Google Scholar, Sept 2021). Through QIIME forums and educational workshops, the development team regularly interacts with users, and these interactions have been vital in helping refine the platform and prioritize updates. For example, QIIME 1 users, many of whom were not trained data scientists, were having trouble understanding and reliably reporting the often-complex bioinformatics workflows. This made replicating their research difficult, and the lack of automated logging of all the steps in the workflow often was a barrier to providing technical support to users. The QIIME 2 developers addressed this need by creating a decentralized provenance tracking system. This system automatically logs each step in an analysis and stores this information as part of its results. This automated logging helped take the guesswork out of QIIME 2 workflows and allowed for fully reproducible cancer microbiome bioinformatics.
Similarly, through interactions with the QIIME 1 user community, the developers learned that users were having difficulty with the command line interface. As a result, the QIIME 2 design enabled access to the same methods through different interface types, including a command line interface and a programmatic interface. As of 2021, QIIME 2 also supports Galaxy, a public platform for processing large data sets in a powerful online infrastructure. Through Galaxy’s popular graphical user interface, researchers now have full access to QIIME 2’s functionality. This new functionality, supported by ITCR, means that cancer researchers can now use QIIME 2 without the need for a command line interface or programming experience.
Finally, demand from the QIIME 1 user community for new methods (as well as from microbiome bioinformatics tool developers who wanted to have their methods accessible through QIIME 1) led the development team to build QIIME 2 based on a plug-in architecture. Now, all bioinformatics functionality in QIIME is implemented in plug-ins, and anyone can develop and disseminate a QIIME 2 plug-in.
QIIME 2 and Cancer Research
QIIME 2 is proving especially relevant for cancer research. For example, several recent studies show that the microbial organisms present in a person or model organism’s gut can directly impact immunotherapy outcomes. A phase I clinical trial by Baruch and colleagues recently found that tumor size was reduced following immunotherapy in 3 of 10 patients who received a fecal microbial transplant (FMT) prior to treatment and who previously hadn’t responded to immunotherapy.1 FMT donor material was harvested from people who already had shown a positive response to treatment. The investigators used QIIME 2 to examine the composition of the donor microbiomes as well as the efficiency of the transplant in the recipients’ gut. Following FMT, the three responding patients’ microbiomes clearly changed to resemble the microbiome of the donor.
In another study, researchers looked at infections linked to treatment for Acute Lymphoblastic Leukemia.2 Using QIIME 2 to analyze 16S data, the investigators found that people who experienced infectious complications in the first 6 months after treatment had significantly lower gut microbiome richness (i.e., reduced variety of microbes) compared with those who didn’t have infections. These studies1,2 show how QIIME 2 is helping us better understand the microbiome’s impact on cancer treatment.
Overall, these studies illustrate the versatility of QIIME 2’s modular design. QIIME 2 users may run complete analysis workflows, as in the two treatment studies, or users can integrate it as a component in other customized workflows.
Next Steps
Interestingly, the two original research studies1,2 described above generated data using both 16S and shotgun metagenomic sequencing. Blending these analyses is becoming increasingly common. It enables researchers, for example, to use lower-cost marker gene sequencing to look at a large number of samples, and then to conduct in-depth analysis of select samples through the higher-cost shotgun sequencing.
A core aim of the QIIME 2 ITCR project is to support the use of multiple types of data to examine the microbiome. Developers around the world are currently working on plug-ins for analyzing shotgun metagenomics data, as well as metabolomic, metatranscriptomic, and metaproteomic data. Ultimately, QIIME 2 will support full integration of these different data types to better understand the human microbiome as it relates to cancer.
That work is already underway, as Bokulich and colleagues built Random Forest models from combined microbiome, immunoproteome, and metabolome data to predict characteristics in the host related to cervical carcinogenesis.3 By integrating microbiome and metabolome data, the researchers were able to predict cancer biomarker levels, suggesting that the host’s microbiome is linked to genital inflammation, which, in turn, may increase the risk for cervical cancer development. Integrating microbiome multi-omics data likely will help us better understand the relationship between the microbiome and cancer. Some of this method-development work will be performed as part of the QIIME 2 ITCR project.
In addition to developing better multi-omics support in QIIME 2, other new features are in progress that will be particularly useful for cancer researchers. For example, plans are underway to improve support for FMT studies, including contextualized analysis of donor microbiomes and techniques to quantify and visualize microbiome engraftment after FMT. This includes offering clinician-accessible reports and statistics summarizing the efficacy of a FMT. Standardizing the approaches for evaluating microbiome engraftment will help improve our ability to match donors and recipients, boosting the odds of successful engraftment and positive clinical outcomes.
As noted above, QIIME 2 development is driven by community input. The best way to join the QIIME 2 community is by registering for a free account on the QIIME 2 Forum. Continued feedback will be vital, both in further refining QIIME 2 and in advancing our understanding of the complex link between the microbiome and cancer. We hope you’ll get involved!
Resources
To learn more about QIIME 2, you can read the documentation or attend a 5-day, hands-on workshop in collaboration with NIH’s Foundation for Advanced Education in the Sciences in early 2022. Registration is open for this event.
References
- Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S, Bloch N, Rotin D, Anafi L, Avivi C, Melnichenko J, Steinberg-Silman Y, Mamtani R, Harati H, Asher N, Shapira-Frommer R, Brosh-Nissimov T, Eshet Y, Ben-Simon S, Ziv O, Khan MAW, Amit M, Ajami NJ, Barshack I, Schachter J, Wargo JA, Koren O, Markel G, Boursi B. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021 Feb 5;371(6529):602-609. doi: 10.1126/science.abb5920. Epub 2020 Dec 10. PMID: 33303685.
- Nearing JT, Connors J, Whitehouse S, Van Limbergen J, Macdonald T, Kulkarni K, Langille MGI. Infectious Complications Are Associated With Alterations in the Gut Microbiome in Pediatric Patients With Acute Lymphoblastic Leukemia. Front Cell Infect Microbiol. 2019 Feb 19;9:28. doi: 10.3389/fcimb.2019.00028. PMID: 30838178; PMCID: PMC6389711.
- Bokulich NA, Laniewski P, Chase DM, Caporaso JG, Herbst-Kralovetz M.M. Integration of multi-omics data improves prediction of cervicovaginal microenvironment in cervical cancer. medRxiv 2020.08.27.20183426; doi:https://doi.org/10.1101/2020.08.27.20183426
Leave a Reply
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (40)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (25)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (13)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

H Nakibapher Jones Shangpliang on October 16, 2021 at 11:43 a.m.