Cancer Data Science Pulse
Machine Learning and Computer Vision Offer a New Way of Looking at Cancer
Copious amounts of data are being collected each day, and researchers are seeking more efficient ways to take advantage of these deep reservoirs of information. One way is through the use of artificial intelligence (AI) and machine-assisted technology.
Once a sci-fi fantasy, AI is now a part of our everyday lives. Virtual assistants, such as Siri and Alexa, can place grocery orders and control other “smart” devices throughout our homes. Self-driving cars are a reality and robots can deliver carry-out orders. Not surprisingly, this robust technology has captured the attention of biomedical scientists as we seek to advance our abilities to understand, diagnose, and treat diseases; and improve public health.
The potential for AI to reshape the health care field and to effectively bring precision medicine into routine clinical care is seemingly boundless. Here we look at a branch of AI called Computer Vision (CV). This technology shifts tasks, such as interpreting an MRI or reading a histology slide—which have long been associated with humans—to an automated machine-based system. It’s giving us a new way to understand and analyze images and has broad implications, particularly for the field of cancer research.
This blog examines a technique described by Dr. James Zou of Stanford University in a recent Center for Biomedical Informatics and Information Technology (CBIIT) Data Science Seminar. He showed how this technology is helping to create a new data-driven “language of morphology” that allows us to be more precise in our study of histological images. In much the same way that computers help propel self-driving cars along busy roadways, this technology offers a faster, less-subjective method for assessing disease. Moreover, with the right training, the use of CV in clinical medicine is proving to be at least as accurate as the human eye.
Image Recognition
Dr. Zou and his team initially used CV as a way to supplement or even replace humans in interpreting echocardiograms (also called ECGs or EKGs). ECGs are the chief diagnostic tool for monitoring heart function; however, interpreting them can be a time-consuming, repetitive, and subjective task. Waiting for ECG assessments also can be a bottleneck, making this an ideal candidate for automation.
Dr. Zou and his colleagues developed an algorithm that assessed cardiac output by measuring the heart’s chamber area. The algorithm assigned a “temporal segmentation” as the model’s key focus, meaning it looked at how the ventricle size differed across time. This then was evaluated on a beat-by-beat basis to arrive at the final output (or ejection fraction). From that, the researchers could predict with high accuracy the patient’s risk for heart failure. The model also allowed them to predict a patient’s risk for related kidney and liver failure.
The model was tested across many different hospital settings with diverse data qualities and various workflows—all of which helped to “train” the system to increase its accuracy and robustness. In the end, they were able to develop an incredibly precise heart algorithm.
Will It Work for Cancer?
The ability to use CV to follow an event over time was key to the success of the ECG model. The researchers next turned their attention to histology. They hypothesized that they could use computers to automatically track cells and cell clusters and to predict not only how those cells looked but how they would behave. Such cell characterization offers a “learned representation” or “deep cellular phenotyping.”
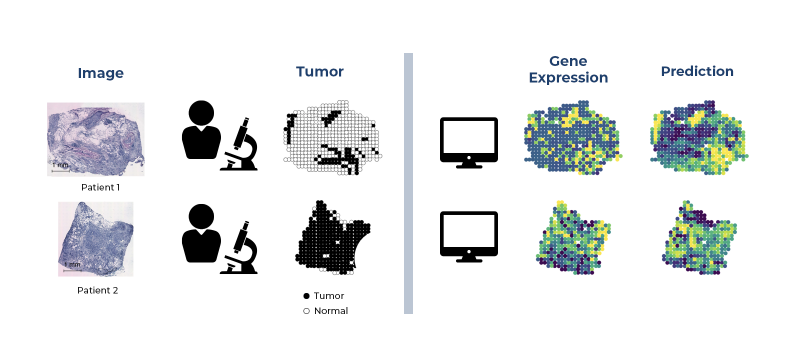
In a typical clinical workflow, histological images and tumor cells are siphoned out to determine what is “normal” and what is “diseased.” Often, a parallel workflow is underway in which the tumor undergoes genomic analysis through a process known as RNA-sequencing (RNA-seq). The results help determine how invasive the tumor is and provide a basic understanding of the genes that might be underlying the disease.
Using RNA-seq gives an abundance of data from one cell. Unfortunately, this type of genetic analysis loses some precision, as it only gives you one snapshot in time, and single-cell analysis doesn’t provide detail beyond the surface. For example, it doesn’t allow researchers to segment structures (i.e., organelles) within a cell or follow individual cells to see how they might interact with their neighboring cells or tissues.
By dovetailing two technologies (histology and spatial genomics), Dr. Zou and his colleagues hoped to create a more finely-grained detail of a typical histological image. Their ultimate goal was to “train” a system to see what the human eye sees, interpret the image, and assess how morphology will progress. Most importantly, they were hoping to create a technique that would be consistent across images and applicable to a variety of settings.
According to Dr. Zou, their team used probes that were about 100 microns each. They overlapped tissue samples across a chip and then attached a barcode that encodes the probe’s physical location to the genes of interest to arrive at a unique molecular identifier (UMI). UMIs enable us to detect genes and other gene products that are likely to be involved in cancer and help to weed out errors that might have been introduced in collecting and processing the sample.
Using this method, they identified variation in gene expression and then trained and validated an algorithm to link the variation in gene expression to the tissue morphology seen in the sample. This gave them a way to translate the image to a particular transcriptome profile, ultimately leading to a deeper understanding of the molecular markers associated with the histological image.
Their method had some key advantages. It allowed them to input the expression of more than 100 genes at a very small (100-micron) resolution. This gives a much more detailed representation than standard histological analysis. To put this into perspective, a standard analysis looks at the entire biopsy slide (often centimeters in size) to arrive at an aggregated assessment. The results often mask fine-grained tissue differences. CV, on the other hand, enables clinicians to “see” the molecular changes in different parts of the biopsied tissue in real time and without having to perform expensive genomics measurements (see Figure 1).
Future Applications
Clearly, there’s great potential in integrating multi-omics and clinical data to better inform patient care and advance precision medicine. As noted by Dr. Zou, “Morpho-dynamic changes ‘come to life’ and you can see how the cells cluster according to their movement.” Seeing how cells move gives you greater insight into how cancer progresses. Coupling this with the transcriptome of cells gives you expression dynamics as well.
This technology could be applied to CRISPR and could enable us to better identify safe targets for genome edits, resulting in more targeted treatment for people with cancer. We’ll have a better idea of how a single cell or small group of cells respond to cancer as well as how the cancer may spread and how to stop or reverse the progression of disease.
The technology still faces some significant hurdles. It’s time-consuming and costly to “train” systems and to assign the correct algorithms. Greater harmonization and aggregation of existing data are needed to make models more robust. Also, as of now, this technology requires highly specialized knowledge. So the next steps will be to translate this technique into the busy clinicians’ workflows to make it efficient and realistic for everyday use. Batch effects such as time of preparation, investigator, type of machine, and more can impact findings. Finding a way to reduce this “technical noise” is vital if this technique is to be used in a variety of settings.
Nevertheless, CV is a promising technique that bears watching as we continue to refine our bioinformatic methods to better understand cancer and similar complex diseases. There is no doubt that AI-related algorithms will create enduring benefits to the health care field, especially precision medicine. It also is critically important to acknowledge that the success of these technologies hinges on forging strong collaborations among all the health care sectors, as well as garnering support from the patients themselves.
Sources
Ouyang, D., He, B., Ghorbani, A., Yuan, N., Ebinger, J., Langlotz, C., Heidenrich, P., Harrington, R., Liang, D., Ashley, E., Zou, J. Video-based AI for beat-to-beat assessment of cardiac function. Nature (2020).
He, B., Bergenstrahle, L., Stenbeck, L., Abid, A., Andersson, A., Borg, A., Maaskola, J., Lundeberg, J., Zou, J. Integrating spatial gene expression and breast tumour morphology with deep learning. Nature Biomedical Engineering (2020).
Zou, J., Huss, M., Abid, A., Mohammadi, P., Torkamani, A., Telenti, A. A primer on deep learning in genetics. Nature Genetics (2019) 51: 12 January; 12–18.
Leave a Reply
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (40)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (25)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (13)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

Coyotetools on November 19, 2022 at 02:37 a.m.