Cancer Data Science Pulse
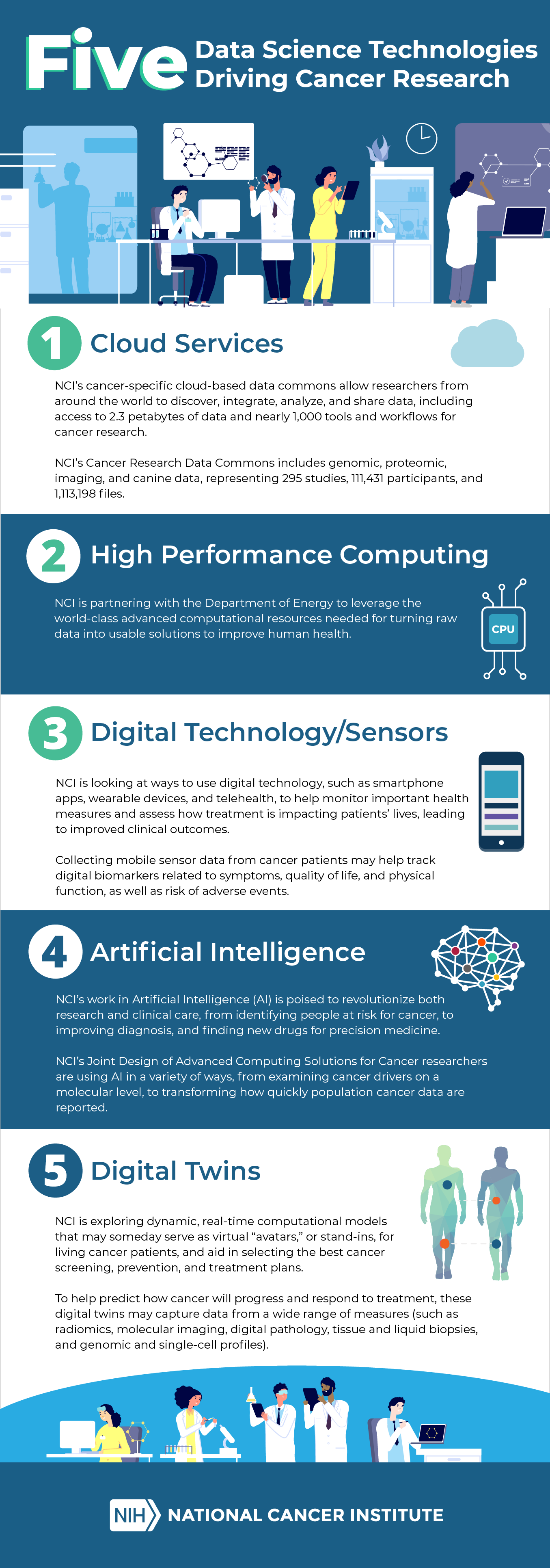
Five Data Science Technologies Driving Cancer Research
To commemorate the National Cancer Act’s 50th anniversary, we’ve pulled together Five Data Science Technologies poised to make a difference in how cancer is diagnosed, treated, and prevented.
Scroll through the infographic below and see if you agree. Some tools are still in their infancy and not quite ready for prime time, whereas others already are revolutionizing research—helping to reveal the underpinnings of cancer and guiding highly tailored precision medicine for more effective treatments.
We’d love to hear your ideas too! Leave your feedback about these technologies or others you're using in the comment form below.
Leave a Reply
https://www.scr.com.au/services/
<a href=”https://skillslash.com/data-science-course-in-dehradun ”> Data science course in Dehradun</a>
Categories
- Data Sharing (66)
- Informatics Tools (42)
- Training (40)
- Precision Medicine (36)
- Data Standards (36)
- Genomics (36)
- Data Commons (34)
- Data Sets (27)
- Machine Learning (25)
- Artificial Intelligence (25)
- Seminar Series (22)
- Leadership Updates (14)
- Imaging (13)
- Policy (10)
- High-Performance Computing (HPC) (9)
- Jobs & Fellowships (7)
- Semantics (6)
- Funding (6)
- Proteomics (5)
- Information Technology (4)
- Awards & Recognition (3)
- Publications (2)
- Request for Information (2)
- Childhood Cancer Data Initiative (1)

wellinnston on November 26, 2021 at 04:41 a.m.
Researchers currently use this data to analyze the disease on three levels:
Cellular - Researchers typically seek for particular patterns in the data to reveal genetic biomarkers that can assist us with predicting tumor mutations and drug therapy.
Patient-Researchers can use their tumor and gene type knowledge to determine the greatest treatments for patients based on their medical history and DNA data.
Population – Treatment alternatives for cancer patients vary based on their different lifestyles, regions, and types of cancer.
The Genome Sequence is one of the most frequent ways to study cancer, during which we analyze the DNA sequence of a single, homogeneous or heter
The real challenge with this huge data set is to find the proper tool to store it for a long time, process, analyze and visualize it.
Although exact retention and deletion policies for data sets are not specified, the fact that they exist demands a method for archiving and keeping them indefinitely.
Data Collection/Transport Mechanism: SFTP,Hadoop Discp, Apache NiFi
Processes/Analyze: Apache Hadoop/MapReduce, Apache Hive, Apache Spark in ML Library,R Packages on top of Spark,Vertica, ETL tools like
Visualization: Tableau,Microstrategy,d3js.
Thanks