Copy Number Variation (CNV) Calling: The Basics
If you’re a data scientist who wants to break into genomics research, you’ll need to know about Copy Number Variation—an essential phenomenon that lets you study the genes underlying cancer and other diseases and disorders. Here we share the basics behind this biological concept, including why cancer researchers use CNVs and how you can apply your data science or bioinformatics skills to capture data based on this fundamental approach.
What is “CNV”?
Our ability to sequence or “read” DNA has revolutionized the field of cancer research, but interpreting this genetic information is the key to understanding how form leads to function. Thanks to extensive mapping of existing human genomes (i.e., telomere to telomere), we now have references to use for comparison. Although it’s not foolproof (as there’s so much genetic diversity across our population), this mapping lets us compare genetic material—numbers of copies and their variations (or CNVs)—to draw conclusions about health and disease.
We use reads to compare CNVs to a previously mapped reference set, or section, of the genome. When you “call” a gene, you’re able to see its location (as opposed to other “noise” or non-coding elements that may or may not influence disease).
Using “probes” or data points of short genome sections, we can easily identify similarities and differences between our sample and the reference. The reference set you choose is a critical part of Next Generation Sequencing (NGS) technology. Your reference will help you translate your results. The more closely the reference genome matches the genome you’re studying, the more accurate your results.
Why is CNV Calling Important for Cancer Research?
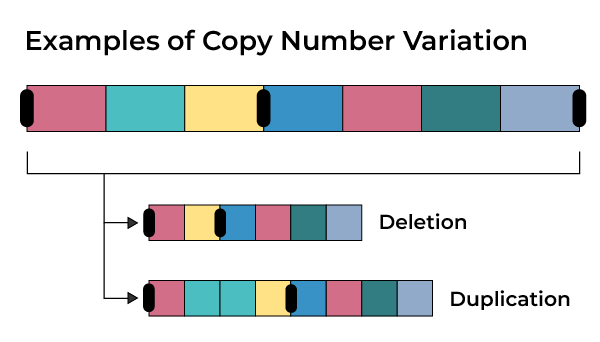
CNVs link to structural variations (such as deletions, duplications, insertions, or other abnormalities), which result in either a loss or gain of DNA. These variations in the genome underlie diseases, such as cancer. See image.
You can use CNV data to look at both germline (i.e., inherited traits and features) or somatic alterations (i.e., changes that arise during a lifetime that are unique to a certain patient or group).
In short, CNVs help us see what’s normal and what’s not (i.e., cancer). Using this approach, we can narrow our search, allowing us to find mutations and alterations that are both drivers of disease and passengers (i.e., they’re passed along without having an impact). It’s these drivers that we can target for potential treatments.
For example, we can target therapies that are more likely to be effective because we know people with certain mutations and alterations are more apt to respond to certain medications. This lets us choose the best treatment for each patient at just the right time. This is the cornerstone of precision medicine.
The potential for this approach is just beginning. Using CNVs, we’re gaining greater insight into the mechanisms behind genetic variations. Someday, CNVs may be a routine part of clinical practice, allowing us to target cancer therapies to each person’s cancer with less risk of toxicity and greater chances of a successful outcome.
What Do I Need to Know?
How Do I Generate CNV Data?
Define what you are searching for and determine how you will conduct your search. For example, are you using CNVs to confirm a cancer diagnosis? Or perhaps you want to use CNVs so you can target medication that is most likely to be effective against a particular cancer. In both of these cases, you likely already know the gene(s) you’re searching for, so the biomarker is clear. You can compare to a reference assay for well-known genes with a stable copy number.
But there’s a lot more you can learn using CNV. Today, you can map variants across cancers to identify genes that help suppress cancer. You also can use CNVs for tracking cancer’s progression—giving you clear markers you can use to identify different stages of disease. And you can use CNV data to predict outcomes for many different cancer types.
- Consider sensitivity and specificity when deciding what to measure. In general, the shorter your testing-window size, and the fewer your probes, the higher the sensitivity and lower the specificity (which could lead to more false positive results). Likewise, a longer window and greater number of probes will give you higher specificity and lower sensitivity. Your sensitivity and specificity also depend on the resolution you choose—whether you’re examining the whole genome (WGS) or whole exome (WES). Resolution hinges on length; that is, are you measuring a section that’s uniform across the region or does it vary in length? With WGS, your coverage tends to be more uniform across the genome, giving you better sensitivity and specificity. On the other hand, when you target select sections, you lose sensitivity and specificity, because you’re looking at a smaller area, which likely includes noncoding regions.
Where Can I Find CNV Data?
- Genomic Data Commons (GDC): Visit NCI’s GDC to access pipelines for analyzing and visualizing CNV data.
What Are Some Examples of Current Algorithms?
Several algorithms exist to help you in interpreting CNVs. For example, in a recent benchmarking study, Dr. Daoud Meerzaman and his colleagues from NCI CBIIT’s Informatics and Data Science Program teamed with other scientists to assess the performance of some popular CNV callers, all of which are good representatives of today’s bioinformatics tools:
- ascatNgs (Allele-Specific Copy number Analysis of Tumors Next Generation Sequencing) helps you work with WGS data in NCI’s GDC platform.
- CNVkit enables you to analyze both WES and WGS data.
- FACETS (Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing) lets you analyze WGS, WES, and targeted (panel) sequencing.
- DRAGEN (Dynamic Read Analysis for GENomics) gives you a scalable tool for identifying variants of all sizes and locations.
- HATCHet (Holistic Allele-Specific Tumor Copy-Number Heterogeneity) helps you analyze variants and duplications jointly across tumor samples.
- Control-FREEC enables you to analyze WGS data. When using with WES data, you will need to include a matched normal sample.
- CNVFam is optimized for comparing genes in families on the sequencing platform, Illumina.
You want the tool you select to give you results that are both consistent and reproducible when applied to WES and WGS data sets. The following can impact your tool’s accuracy:
- The sequencing platform (e.g., Illumina is the platform for DRAGEN and CNVfam)
- The sample preparation (i.e., fresh or formalin-fixed paraffin-embedded, also known as “FFPE”)
- The amount of sequencing coverage (i.e., ranging from 10 to 300X coverage)
Tips for Working with Genomic Data
- The number of sets of chromosomes (i.e., “ploidy”) matters and has significant downstream effects. That is, your results using a single copy of human genome, or “haploid” (i.e., double-stranded DNA), differs from a “diploid” (i.e., two sets of double-stranded DNA). An abnormal number of chromosomes, called “aneuploidy,” is a major contributor to cancer’s development and progression. When comparing caller tools, you’ll find the greatest variation in CNV calls is from genome ploidy.
- Non-analytical factors also can influence your calling results, including whether you’re using FFPE vs. frozen samples. Other factors that impact your results include the purity of your DNA input and the library prep protocol you use to convert your genomic DNA sample for sequencing.
- It’s important to use diverse approaches, rather than relying on one standard tool. For the most precise results, you should consider using multiple CNV calling tools.
NCI CNV Resources and Initiatives
Resources and Tools
- Division of Cancer Epidemiology and Genetics (DCEG): Search these statistical and computational tools featured in DCEG epidemiological and laboratory studies.
- Circle plots for visualizing multi-omics data: Learn how to use visualize your CNV data results.
Publications
Want to know more about CNV analysis? Check out these publications from CBIIT staff and NCI-funded researchers.
Evaluation of Somatic Copy Number Variation Detection by NGS Technologies and Bioinformatics Tools on a Hyper‑Diploid Cancer Genome. Genome Biology, 2024. | Review this study that benchmarks CNV calling performance in six common tools based on their detection accuracy, sensitivity, and reproducibility.
Structural Variant Analysis of a Cancer Reference Cell Line Sample Using Multiple Sequencing Technologies. Genome Biology, 2022. | See how researchers developed a consensus structural variants (SV) call set for benchmarking and developing SV detection methods and algorithms.
Computational Methods for Detecting Copy Number Variations in Cancer Genome Using Next Generation Sequencing: Principles and Challenges. Ocotarget, 2013. | Review common tools used in NGS-based cancer CNV studies, as well as things to keep in mind when analyzing NGS data.