News
CPTAC Investigators Establish Protein Level Prediction Model Using Crowdsourcing
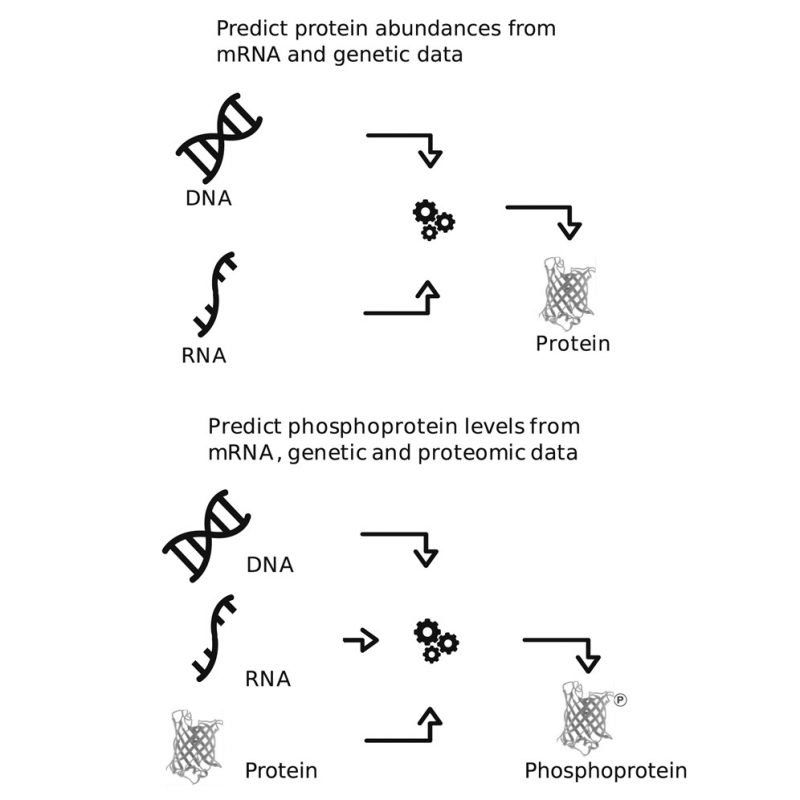
Clinical Proteomic Tumor Analysis Consortium (CPTAC) researchers shared the results of their first crowd-sourced NCI-CPTAC DREAM Proteogenomic Challenge. In this community-based, collaborative contest, participants developed computational algorithms to predict protein abundance and phosphoprotein levels.
Using the power of crowdsourcing, top-performers were able to shed more light on transcriptional and translational control of the proteome and reveal factors that influenced protein predictability. Models from the DREAM Proteogenomic Challenge winners and the DREAM Challenge predictions have been made publicly available to the bioinformatic and cancer research community for use and further development.
For more information about the contest and its results, read the full article on NCI's Center for Strategic Scientific Initiatives website.